plotnine-gallery-aez

Figure 1
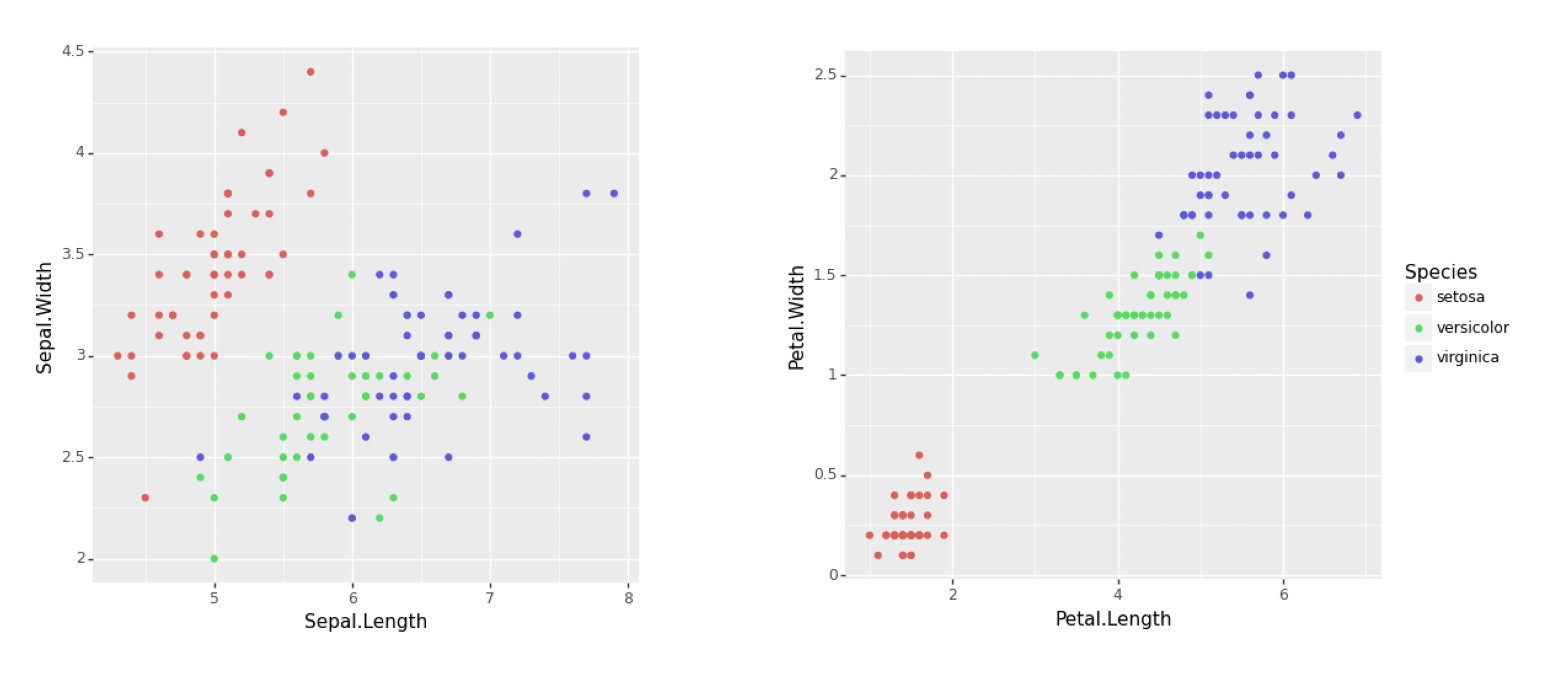
Keywords: combining plots
import matplotlib.pyplot as plt import matplotlib.image as mpimg import matplotlib as matplotlib matplotlib.use('QtAgg') import pandas as pd import plotnine as p9 from plotnine import ggplot, geom_point, aes, theme iris = pd.read_csv("iris.csv") plot1 = (ggplot() + geom_point(data = iris, mapping = aes(x = "Sepal.Length", y = "Sepal.Width", colour = "Species")) + theme(legend_position = "none")) plot2 = (ggplot() + geom_point(data = iris, mapping = aes(x = "Petal.Length", y = "Petal.Width", colour = "Species"))) plot1.save("plot1.png", width=5, height=5) plot2.save("plot2.png", width=6, height=5) img1 = mpimg.imread('plot1.png') img2 = mpimg.imread('plot2.png') fig, axs = plt.subplots(1, 2, gridspec_kw={'width_ratios': [1, 1.2]}) axs[0].imshow(img1) axs[0].axis('off') axs[1].imshow(img2) axs[1].axis('off') plt.savefig("fig01.png", dpi=300, bbox_inches='tight')
Figure 2
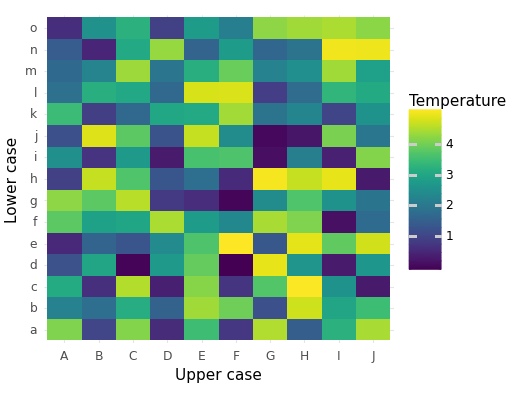
Keywords: heatmap, matrix, tile geometry
import matplotlib.pyplot as plt import matplotlib.image as mpimg import matplotlib as matplotlib matplotlib.use('QtAgg') from plotnine import ggplot, aes, geom_tile, labs, theme_minimal import pandas as pd import numpy as np # Creating the data x = list('ABCDEFGHIJ') y = list('abcdefghijklmno') plot_df = pd.DataFrame( {'x': np.tile(x, len(y)), 'y': np.repeat(y, len(x)), 'z': np.random.uniform(0, 5, len(x) * len(y))}) # Creating the plot example_plot = (ggplot() + geom_tile(data=plot_df, mapping=aes(x='x', y='y', fill='z')) + labs(x='Upper case', y='Lower case', fill='Temperature') + theme_minimal()) fig = example_plot.draw() fig.set_facecolor('white') fig.set_size_inches(13 / 2.54, 10 / 2.54) fig.savefig("fig02.png")
Figure 3
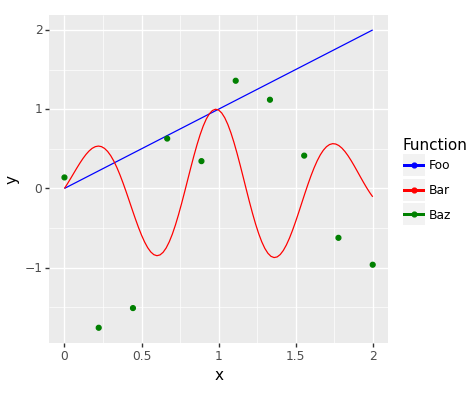
Keywords: function, curve, expression, colours
import matplotlib.pyplot as plt import matplotlib.image as mpimg import matplotlib as matplotlib matplotlib.use('QtAgg') from plotnine import ggplot, aes, stat_function, geom_point, scale_colour_manual import pandas as pd import numpy as np # Function for the first line (identity function) def identity(x): return x # Function for the second line def custom_function(x): return np.exp(-(x - 1)**2) * np.sin(x * 8) # Data for the scatter plot scatter_data = pd.DataFrame({ 'x': np.linspace(0, 2, 10), 'y': np.random.normal(size = 10) }) example_plot = (ggplot(pd.DataFrame({'x': [0, 2]}), aes('x')) + stat_function(fun = identity, geom = 'line', mapping = aes(colour = '"Foo"')) + stat_function(fun = custom_function, geom = 'line', mapping = aes(colour = '"Bar"')) + geom_point(data = scatter_data, mapping = aes(colour = '"Baz"', y = 'y')) + scale_colour_manual(name = "Function", values = {"Foo": "blue", "Bar": "red", "Baz": "green"})) fig = example_plot.draw() fig.set_facecolor('white') fig.set_size_inches(12 / 2.54, 10 / 2.54) fig.savefig("fig03.png")
Figure 4
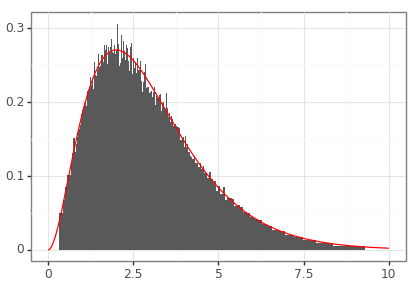
Keywords: credible intervals, confidence intervals, pandas, density
import matplotlib.pyplot as plt import matplotlib.image as mpimg import matplotlib as matplotlib matplotlib.use('QtAgg') from plotnine import * import pandas as pd import numpy as np import scipy.stats def plottable_ci_df(model_ci_df): """ Converts the model_cints table into a dataframe that can be plotted using plot9. Assumes that the model_cints table has the following columns: - prob (as a percentage) - ymin - ymax Returns a dataframe with the following columns: - prob - mass - ymin - ymax """ assert model_ci_df['prob'].dtype == int dd = sorted(model_ci_df.to_dict(orient= 'records'), key=lambda k: k['prob']) left_blocks, right_blocks = [], [] prev_prob = 0 probs = [d['prob'] for d in dd] start_prob = np.array(probs).min() for prob in probs: di = [d for d in dd if d['prob'] == prob][0] left_block_right = (0.5 * ( di['ymin'] + di['ymax'] ) if prob == start_prob else left_blocks[-1]['ymin']) di_left = {'prob': di['prob'], 'mass': 0.5 * (di['prob'] / 100 - prev_prob / 100), 'ymax': left_block_right, 'ymin': di['ymin']} left_blocks.append(di_left) right_block_left = (0.5 * ( di['ymin'] + di['ymax'] ) if prob == start_prob else right_blocks[-1]['ymax']) di_right = {'prob': di['prob'], 'mass': 0.5 * (di['prob'] / 100 - prev_prob / 100), 'ymax': di['ymax'], 'ymin': right_block_left} right_blocks.append(di_right) prev_prob = di['prob'] tmp = pd.concat([pd.DataFrame(left_blocks), pd.DataFrame(right_blocks)]) tmp['xmin'] = 0 tmp['xmax'] = tmp['mass'] / (tmp['ymax'] - tmp['ymin']) return tmp gamma_dist = scipy.stats.gamma(3) # Construct approximate density based on central credible intervals. gamma_rvs = gamma_dist.rvs(size = 100000) cov_prct = np.linspace(1, 99, 99) ci_lower = np.percentile(gamma_rvs, cov_prct / 2) ci_upper = np.percentile(gamma_rvs, 100 - cov_prct / 2) gamma_ci_df = pd.DataFrame({'prob': np.flip(cov_prct).astype(int), 'ymin': ci_lower, 'ymax': ci_upper}) plt_df_1 = plottable_ci_df(gamma_ci_df) # Construct exact density. xs = np.linspace(0, 10, 1000) ys = gamma_dist.pdf(xs) plt_df_2 = pd.DataFrame({'x': xs, 'y': ys}) demo_p9 = (ggplot() + geom_rect(data = plt_df_1, mapping = aes(xmin = 'ymin', xmax = 'ymax', ymin = 'xmin', ymax = 'xmax')) + geom_line(data = plt_df_2, mapping = aes(x = 'x', y = 'y'), color = 'red') + theme_bw() + theme(axis_title = element_blank()) ) demo_p9.save("fig04.png", height = 2.9, width = 4.1)
Figure 5
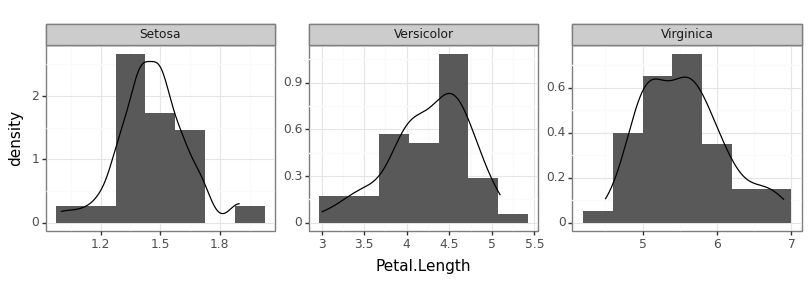
Keywords: facet labeller, title case, density
import matplotlib.pyplot as plt import matplotlib.image as mpimg import matplotlib as matplotlib matplotlib.use("QtAgg") from plotnine import * import pandas as pd import numpy as np import scipy.stats iris = pd.read_csv("iris.csv") def my_labeller(label: str) -> str: return label.title() demo_p9 = ( ggplot(iris, aes(x="Petal.Length", y="..density..")) + geom_histogram(bins=7) + geom_density() + facet_wrap("Species", scales="free", labeller=my_labeller) + theme_bw() ) demo_p9.save("fig05.png", height=2.9, width=8.1)