matplotlib gallery
Figure 1
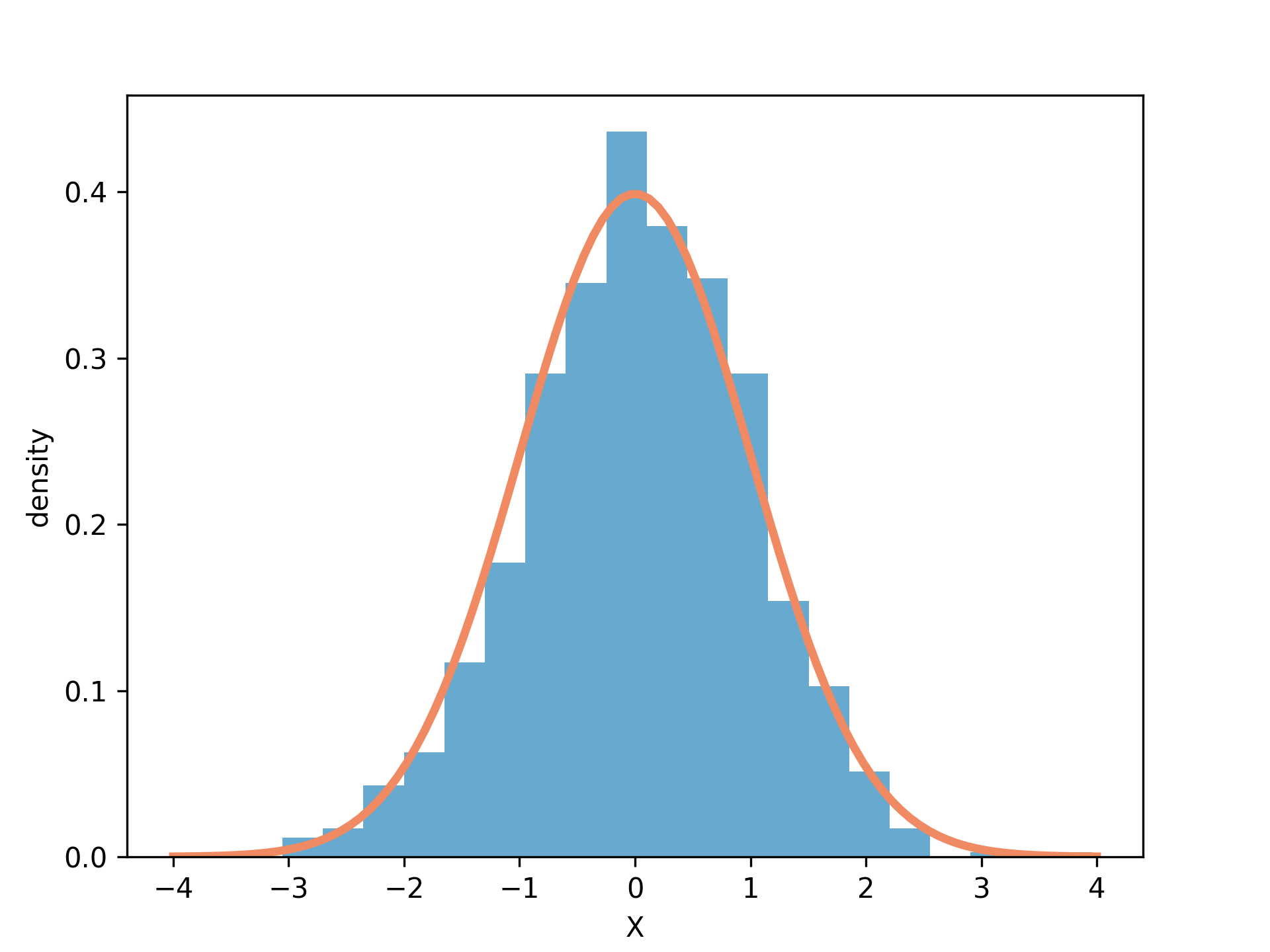
import matplotlib.pyplot as plt import numpy as np np.random.seed(1) x = np.random.randn(1000) x_mesh = np.linspace(start = -4, stop = 4, num = 100) y = np.exp(-0.5 * x_mesh ** 2) / np.sqrt(2 * np.pi) fig = plt.figure() ax = fig.add_axes([0.1,0.1,0.8,0.8]) ax.hist(x, bins = 20, density=True, color="#67a9cf") ax.plot(x_mesh, y, color="#ef8a62", linewidth=3) ax.set_xlabel("X") ax.set_ylabel("density") fig.savefig("fig01.png", dpi=300)
Figure 2
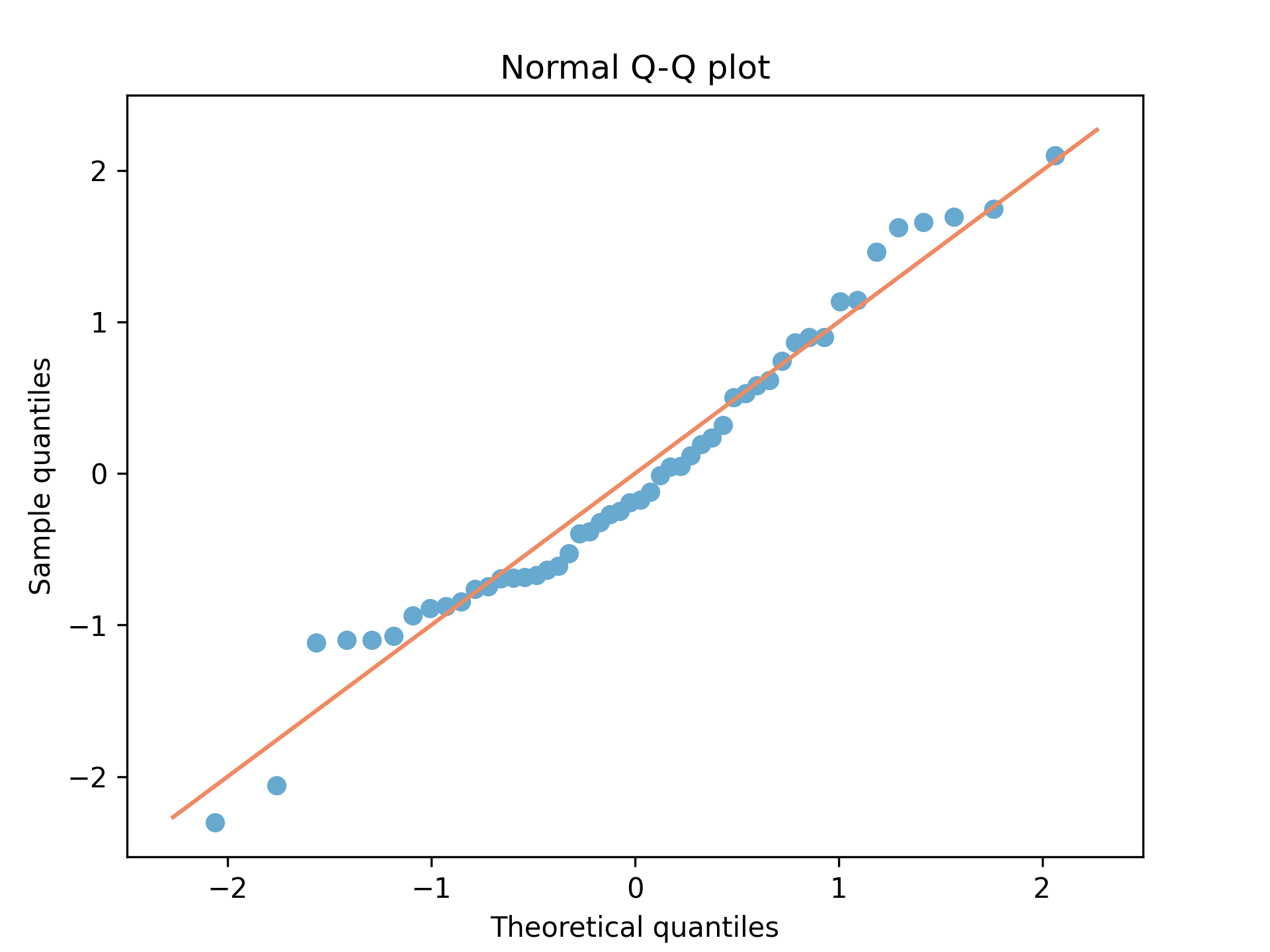
import matplotlib.pyplot as plt import numpy as np import scipy.stats as stats np.random.seed(1) num_points = 50 x = np.random.randn(num_points) x.sort() unit_mesh = np.linspace( start = num_points / (num_points + 1), stop = 1 / (num_points + 1), num = num_points) quantile_vals = stats.norm.isf(unit_mesh) line_lims = [min(quantile_vals) * 1.1,max(quantile_vals) * 1.1] fig = plt.figure() ax = fig.add_axes([0.1,0.1,0.8,0.8]) ax.scatter(quantile_vals, x, color="#67a9cf") ax.plot(line_lims, line_lims, color="#ef8a62") ax.set_title("Normal Q-Q plot") ax.set_xlabel("Theoretical quantiles") ax.set_ylabel("Sample quantiles") fig.savefig("fig02.png", dpi=300)
Figure 3
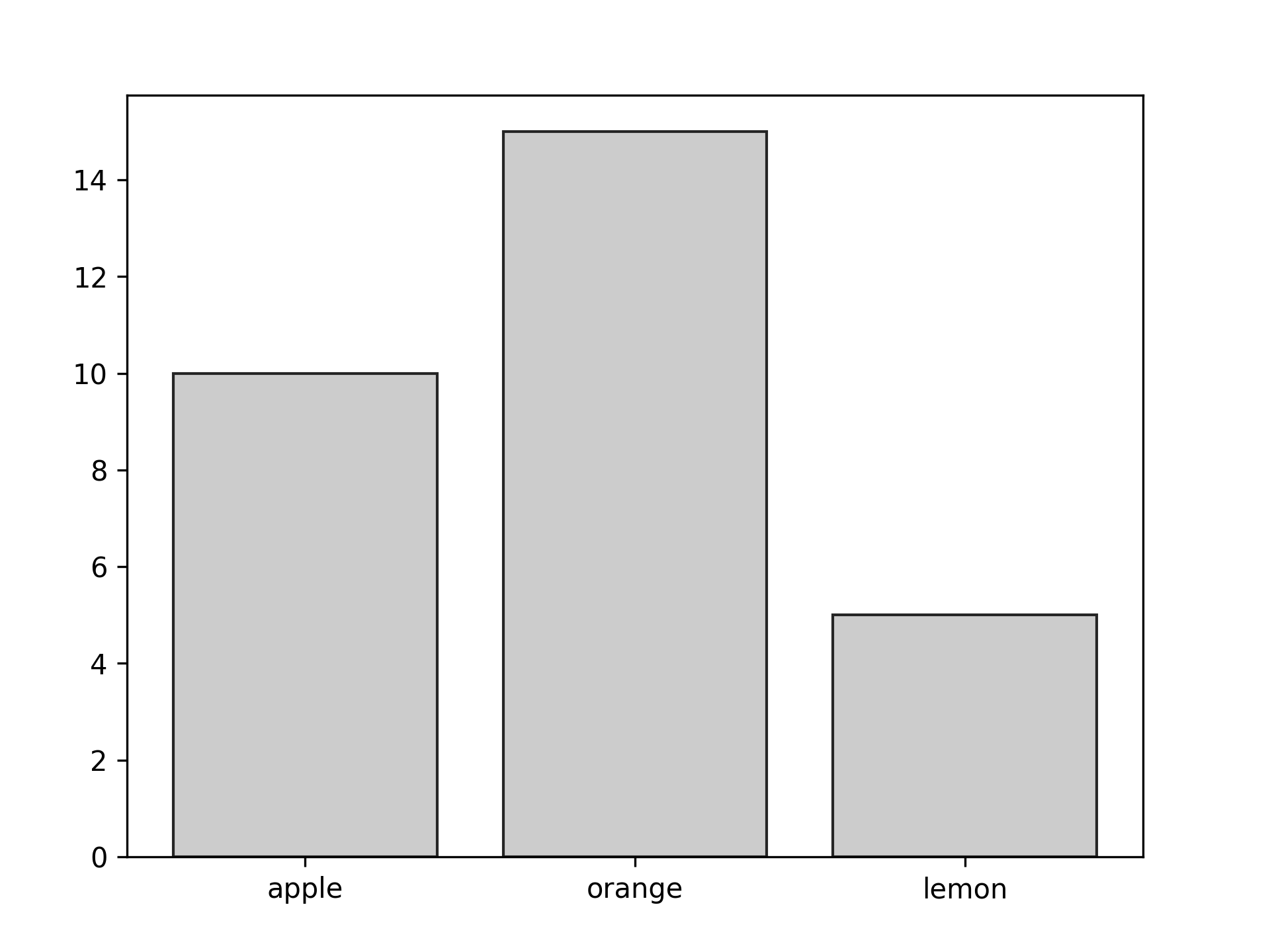
import matplotlib.pyplot as plt data = {'apple': 10, 'orange': 15, 'lemon': 5} names = list(data.keys()) values = list(data.values()) fig = plt.figure() ax = fig.add_axes([0.1,0.1,0.8,0.8]) ax.bar(names, values, color = "#cccccc", edgecolor = "#252525") fig.savefig("fig03.png", dpi=300)
Figure 4
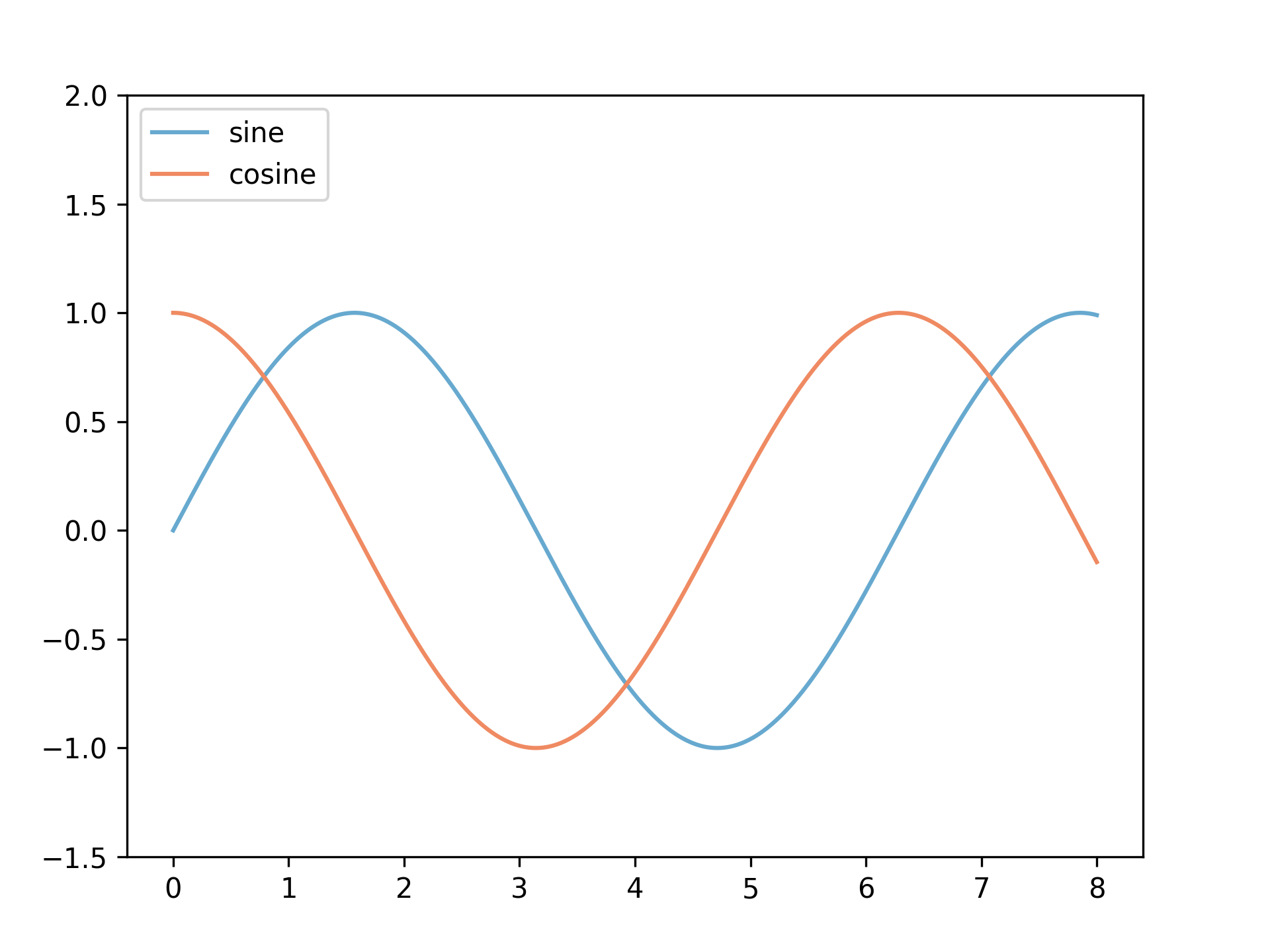
import numpy as np import matplotlib.pyplot as plt x = np.linspace(0, 8, 1000) y1 = np.sin(x) y2 = np.cos(x) fig = plt.figure() ax = fig.add_axes([0.1,0.1,0.8,0.8]) ax.plot(x, y1, color="#67a9cf", label="sine") ax.plot(x, y2, color = "#ef8a62", label="cosine") ax.legend(loc="upper left") ax.set_ylim([-1.5, 2]) fig.savefig("fig04.png", dpi=300)
Figure 5
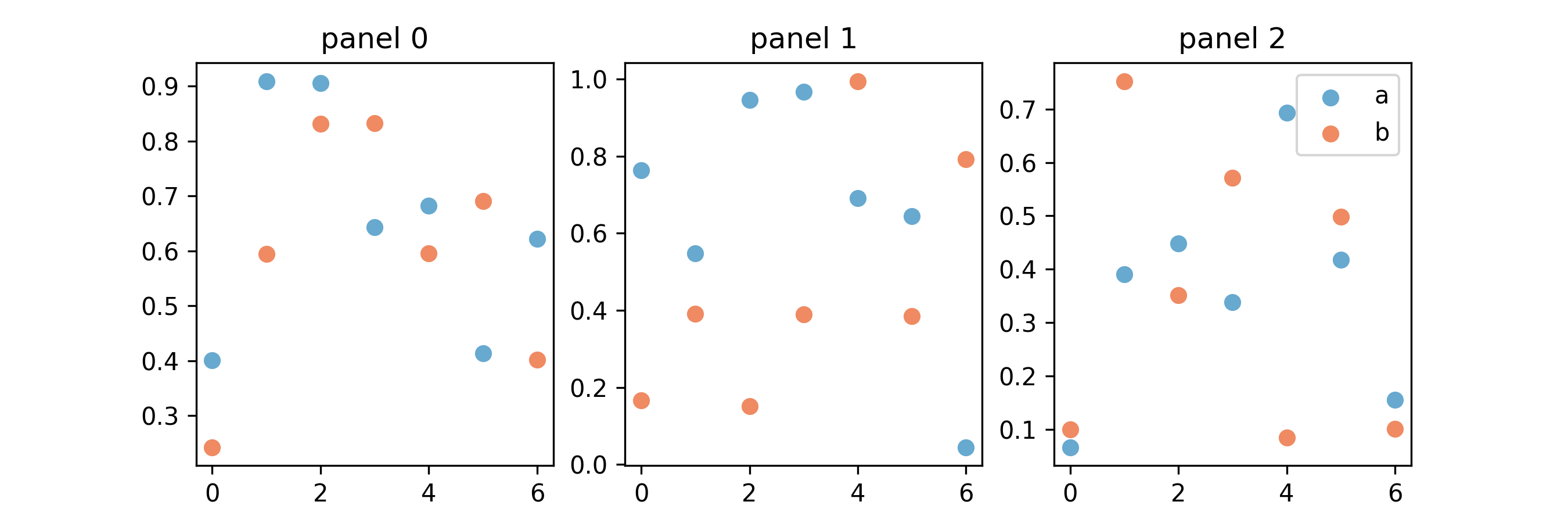
import numpy as np import matplotlib.pyplot as plt fig, axs = plt.subplots(1, 3, figsize=(9, 3)) var_and_col = list(zip(['a', 'b'], ["#67a9cf", "#ef8a62"])) for ix in range(3): for v, c in var_and_col: xs = range(7) ys = np.random.uniform(size=7) axs[ix].set_title("panel {ix}".format(ix=ix)) axs[ix].scatter(xs, ys, edgecolors = c, facecolors = "none", label = v) axs[2].legend(loc = "upper right") fig.savefig("fig05.png", dpi=300)
Figure 6
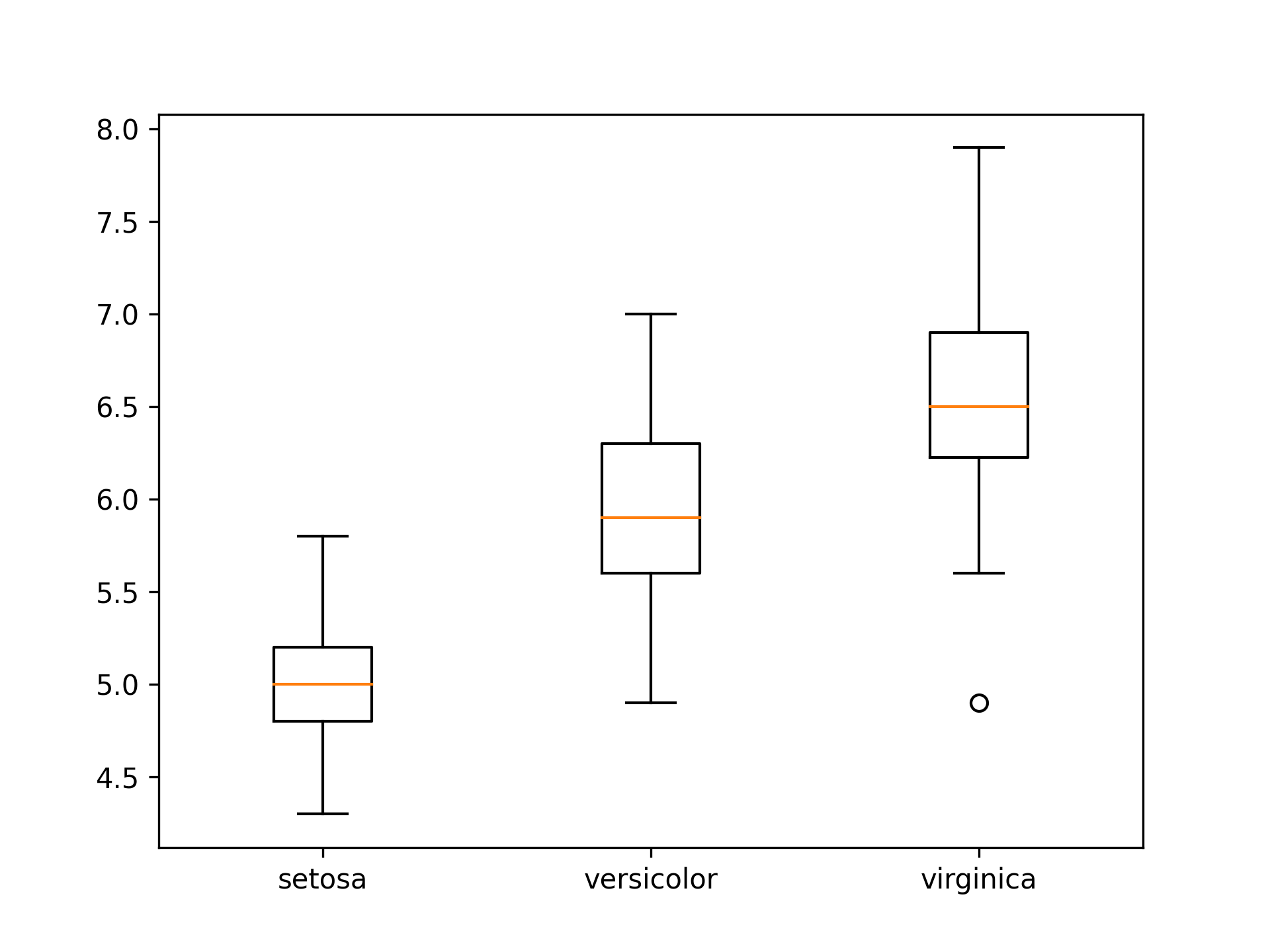
import matplotlib.pyplot as plt import pandas as pd iris = pd.read_csv("iris.csv") unique_species = iris.species.unique() grouped_sepal_lengths = [iris[iris.species == species].sepal_length for species in unique_species] plt.figure() plt.boxplot(x = grouped_sepal_lengths, labels = unique_species) plt.savefig("fig06.png", dpi=300)
Figure 7
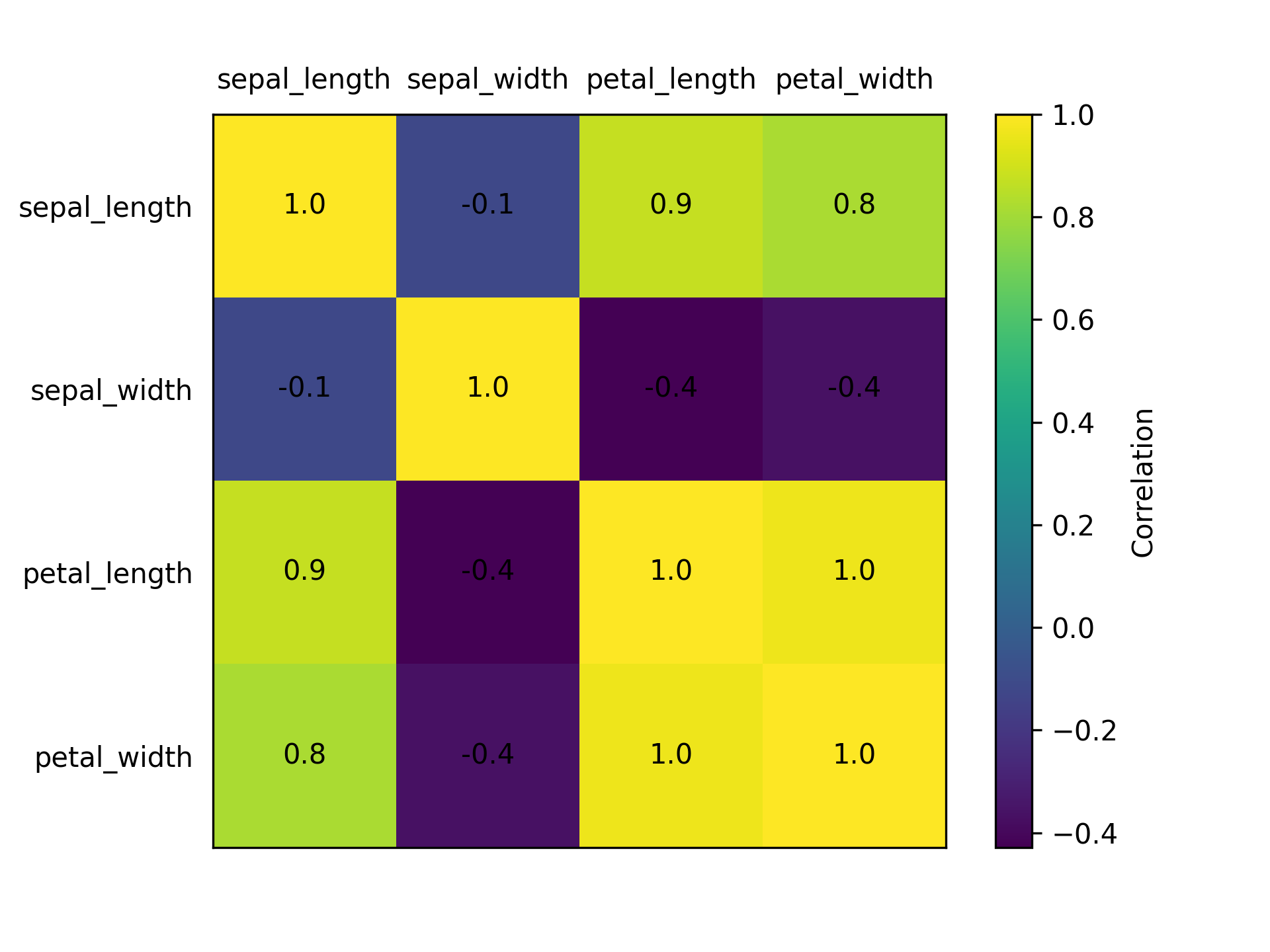
import matplotlib.pyplot as plt import matplotlib as matplotlib import pandas as pd iris = pd.read_csv("iris.csv") numeric_cols = iris.columns.to_list()[0:4] iris_corrs = iris[numeric_cols].corr().to_numpy() fig, ax = plt.subplots() im = ax.imshow(iris_corrs) cbar = ax.figure.colorbar(im) cbar.ax.set_ylabel("Correlation") ax.set_xticks(range(iris_corrs.shape[1])) ax.set_yticks(range(iris_corrs.shape[0])) ax.set_xticklabels(numeric_cols) ax.set_yticklabels(numeric_cols) ax.tick_params(top=False, bottom=False, labeltop=True, labelbottom=False) kw = dict(horizontalalignment="center", verticalalignment="center", color="black") valfmt = matplotlib.ticker.StrMethodFormatter("{x:.1f}") for i in range(iris_corrs.shape[0]): for j in range(iris_corrs.shape[1]): im.axes.text(j, i, valfmt(iris_corrs[i, j], None), **kw) plt.tick_params(left=False) plt.savefig("fig07.png", dpi=300)
Figure 8
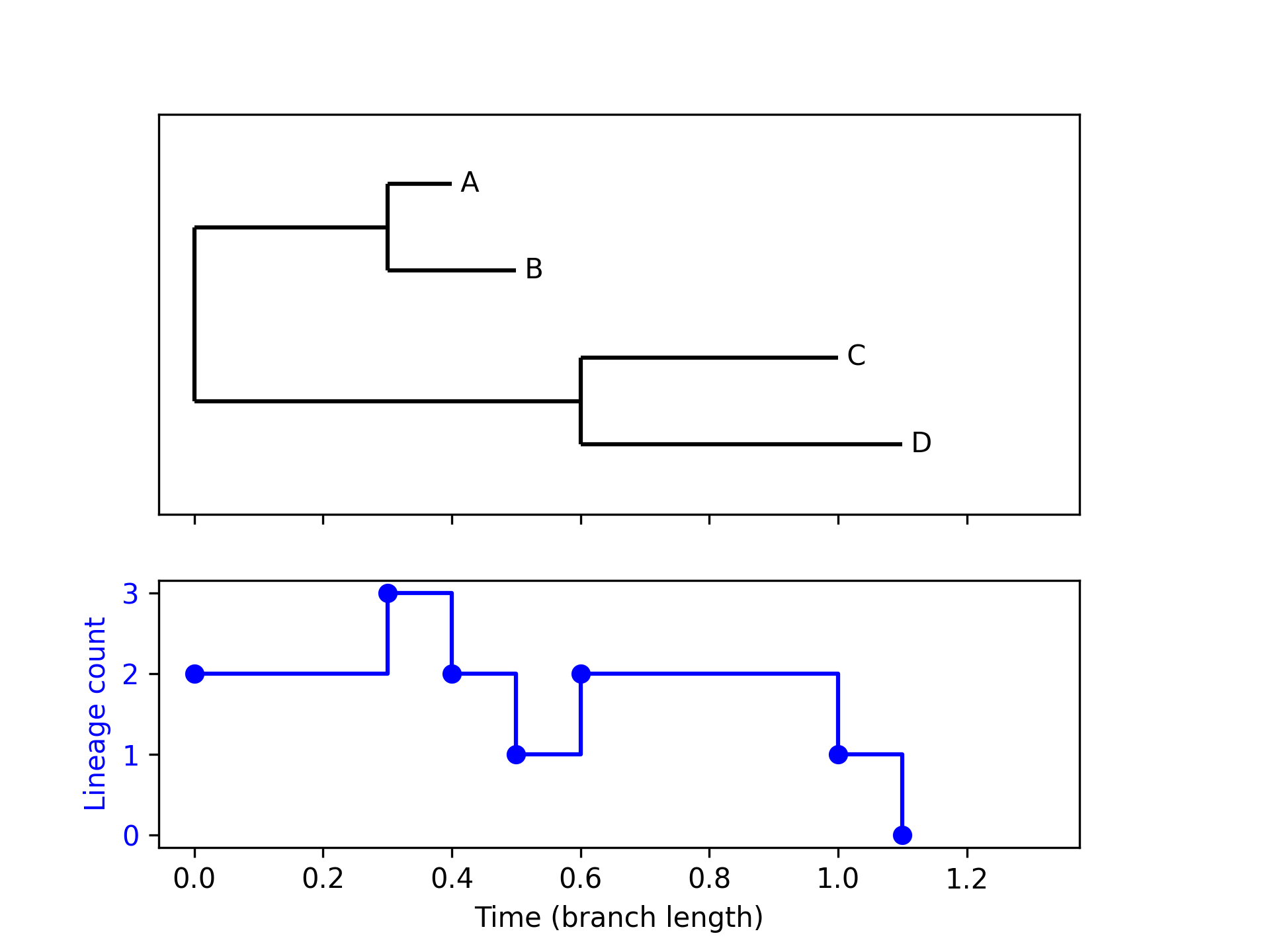
from Bio import Phylo from io import StringIO import matplotlib.pyplot as plt import numpy as np def ltt_data(tree): node_times = [] def traverse(clade, current_time): bl = clade.branch_length if clade.branch_length else 0 if clade.clades: node_times.append(('branch', current_time)) for child in clade.clades: if child.is_terminal(): node_times.append(('tip', current_time + child.branch_length)) else: traverse(child, current_time + child.branch_length) traverse(tree.root, 0) times = [] num_lines = [] curr_ltt = 1 for (node_type, time) in node_times: times.append(time) if node_type == 'branch': curr_ltt += 1 num_lines.append(curr_ltt) else: curr_ltt -= 1 num_lines.append(curr_ltt) return times, num_lines newick_tree = StringIO("((A:0.1, B:0.2):0.3, (C:0.4, D:0.5):0.6);") tree = Phylo.read(newick_tree, "newick") fig, (ax1, ax2) = plt.subplots(2, 1, sharex=True, gridspec_kw={'height_ratios': [3, 2]}) Phylo.draw(tree, do_show = False, axes = ax1) ax1.yaxis.set_ticks([]) ax1.yaxis.set_ticklabels([]) ax1.set_ylabel('') ax1.set_xlabel('') ltt_x, ltt_y = ltt_data(tree) ax2.plot(ltt_x, ltt_y, 'bo') ax2.step(ltt_x, ltt_y, 'b-', where='post', label='Sine Wave') ax2.set_ylabel('Lineage count', color='b') ax2.set_xlabel('Time (branch length)') ax2.tick_params(axis='y', labelcolor='b') plt.subplots_adjust(right=0.85) fig.savefig("fig08.png", dpi=300)
Figure 9
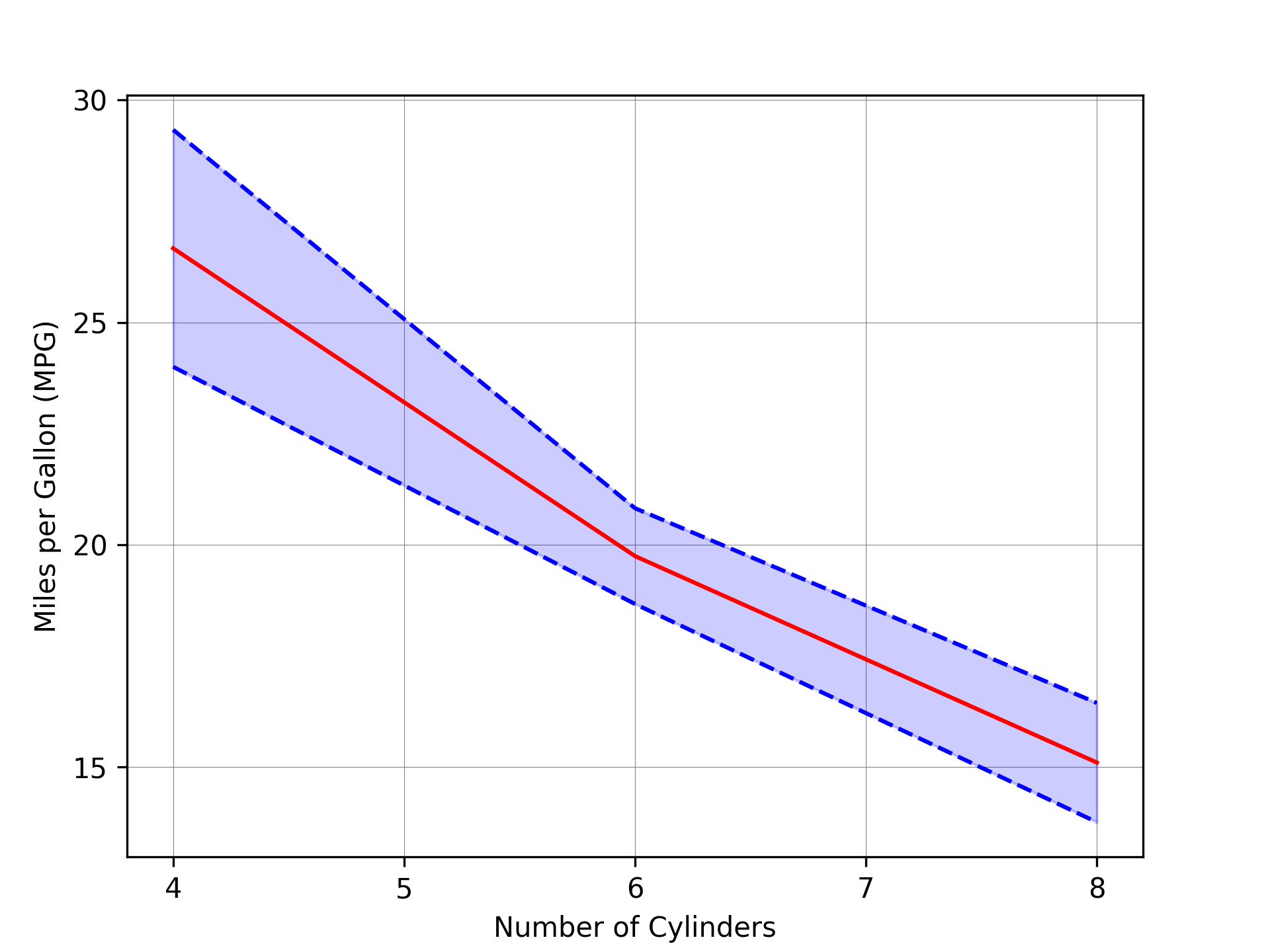
import pandas as pd import numpy as np import matplotlib.pyplot as plt mtcars = pd.read_csv("mtcars.csv") plot_df = ( mtcars.groupby("cyl") .agg( mean_mpg=("mpg", "mean"), sd_mpg=("mpg", "std"), n=("mpg", "size"), # Count the number of occurrences for each group ) .reset_index() ) plot_df["lower_mpg"] = plot_df["mean_mpg"] - 1.96 * plot_df["sd_mpg"] / np.sqrt( plot_df["n"] ) plot_df["upper_mpg"] = plot_df["mean_mpg"] + 1.96 * plot_df["sd_mpg"] / np.sqrt( plot_df["n"] ) fig = plt.figure() ax = fig.add_axes([0.1, 0.1, 0.8, 0.8]) ax.fill_between( plot_df["cyl"], plot_df["lower_mpg"], plot_df["upper_mpg"], color="blue", alpha=0.2 ) ax.plot(plot_df["cyl"], plot_df["lower_mpg"], "--", color="blue") ax.plot(plot_df["cyl"], plot_df["upper_mpg"], "--", color="blue") ax.plot(plot_df["cyl"], plot_df["mean_mpg"], "-", color="red", label="Mean") ax.set_xlabel("Number of Cylinders") ax.set_ylabel("Miles per Gallon (MPG)") ax.set_xticks(range(4, 9)) ax.set_yticks(range(15, 35, 5)) ax.grid(True, which="major", linestyle="-", linewidth=0.25, color="grey", zorder=0) # Ensure the grid lines are in the background by setting the z-order of the plots ax.set_axisbelow(True) fig.savefig("fig09.png", dpi=300)
Figure 10
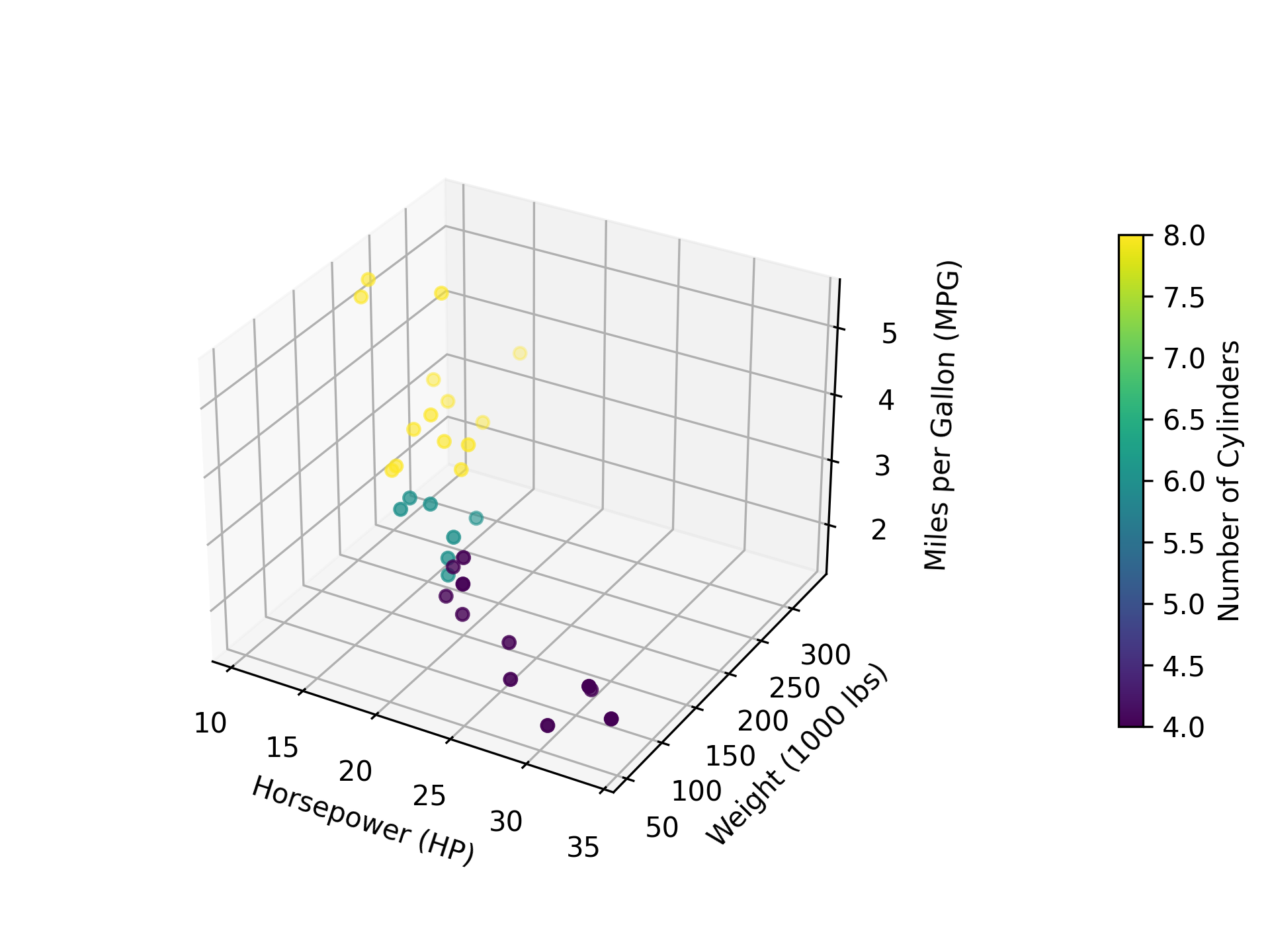
import pandas as pd import numpy as np import matplotlib.pyplot as plt mtcars = pd.read_csv("mtcars.csv") plot_df = mtcars[["mpg", "hp", "wt", "cyl"]] fig = plt.figure() ax = fig.add_subplot(111, projection="3d") scatter = ax.scatter( plot_df["mpg"], plot_df["hp"], plot_df["wt"], c=plot_df["cyl"], cmap="viridis", marker="o", ) colorbar = fig.colorbar(scatter, ax=ax, fraction=0.025, pad=0.25) colorbar.set_label("Number of Cylinders") ax.set_xlabel("Horsepower (HP)") ax.set_ylabel("Weight (1000 lbs)") ax.set_zlabel("Miles per Gallon (MPG)") # For an interactive plot, uncomment the following line. # plt.show() fig.savefig("fig10.png", dpi=300)
Figure 11
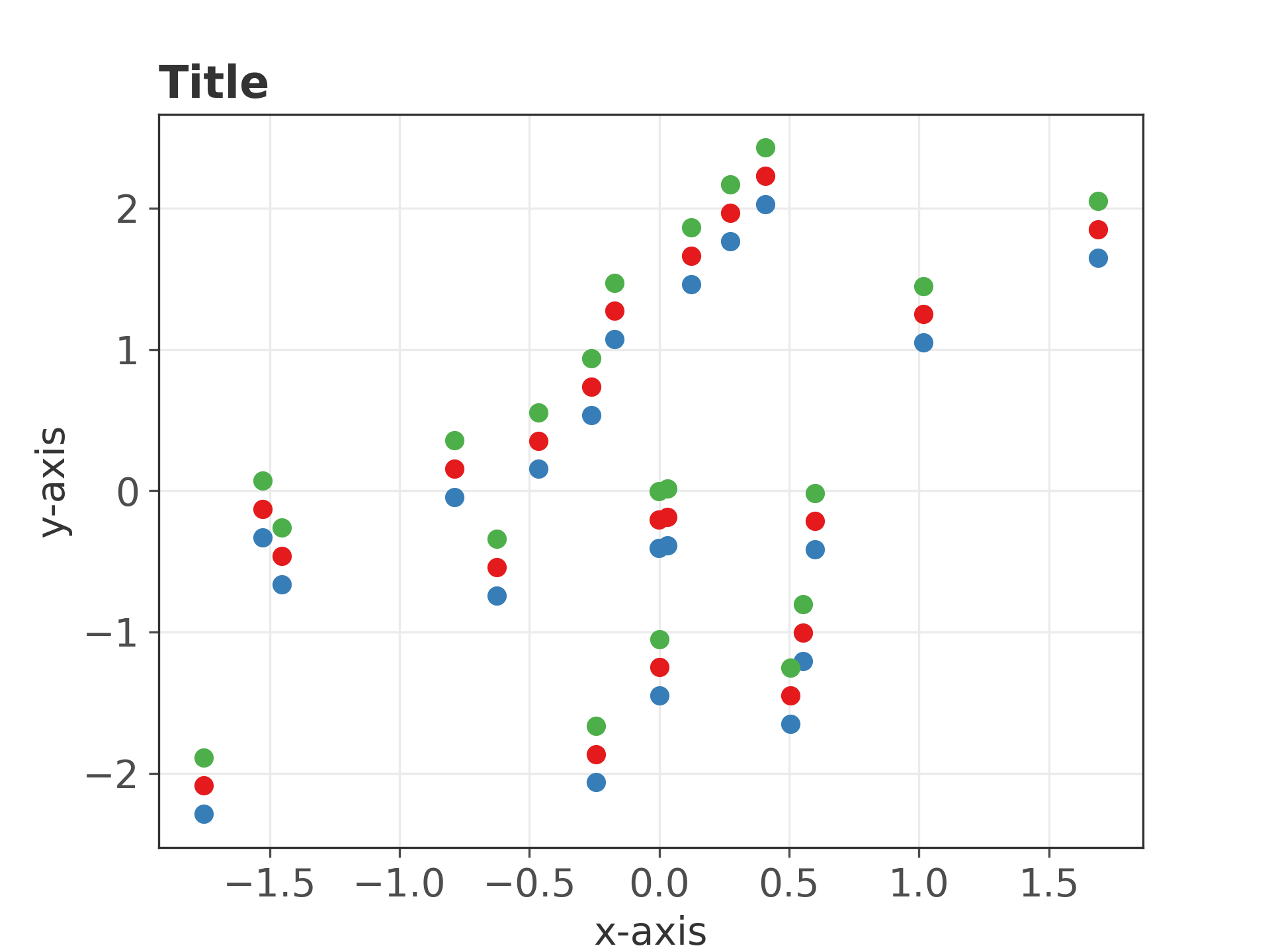
import numpy as np import matplotlib.pyplot as plt np.random.seed(7) x = np.random.randn(20) y = np.random.randn(20) plt.style.use('./aez20250101.mplstyle') plt.scatter(x, y, zorder=5) plt.scatter(x, y+0.2, zorder=5) plt.scatter(x, y+0.4, zorder=5) plt.title('Title') plt.xlabel('x-axis') plt.ylabel('y-axis') plt.savefig("fig11.png", dpi=300)
## =========================================================
## Author: Alexander E. Zarebski
## Date: 2025-01-01
## =========================================================
##
## Matplotlib configuration are currently divided into
## following parts:
##
## - AXES
## - FONT
## - GRID
##
## =========================================================
## ---------------------------------------------------------
## AXIS
## ---------------------------------------------------------
axes.edgecolor: "#333333"
axes.grid: True
axes.labelcolor: "#333333"
axes.titlecolor: "#333333"
axes.titlesize: 16.0
axes.titleweight: "bold"
axes.titlelocation: "left"
axes.prop_cycle: cycler('color', ['377eb8','e41a1c','4daf4a','984ea3','ff7f00'])
xtick.color: "#4d4d4d"
ytick.color: "#4d4d4d"
## ---------------------------------------------------------
## FONT
## ---------------------------------------------------------
font.family: "sans-serif"
font.size: 14.0
## ---------------------------------------------------------
## GRID
## ---------------------------------------------------------
grid.color: "#ebebeb"
Figure 12
import numpy as np import matplotlib.pyplot as plt import squarify np.random.seed(7) x = np.sort(np.exp(np.random.normal(2, 1, 10))) plt.figure() squarify.plot(sizes=x, color=10*["#1b9e77"], label=['A', 'B', 'C', 'D', 'E', 'F', 'G', 'H', 'I', 'J'], pad=True) plt.axis("off") plt.title("Title") plt.savefig("fig12.png", dpi=300)
Figure 13
import numpy as np import matplotlib.pyplot as plt np.random.seed(7) x = np.linspace(0, 10, 200) y_left = np.sin(x) y_right = 100 * np.cos(x) fig, ax_left = plt.subplots() ax_left.plot(x, y_left, color="green") ax_left.set_xlabel("x") ax_left.set_ylabel("sin(x)", color="green") ax_left.tick_params(axis="y", colors="green") left_ticks = [-1.0, -0.5, 0.0, 0.5, 0.6, 0.7, 1.0] ax_left.set_yticks(left_ticks) ax_right = ax_left.twinx() ax_right.plot(x, y_right, color="purple") ax_right.set_ylabel("100 cos(x)", color="purple") ax_right.tick_params(axis="y", colors="purple") # NOTE This is needed to keep the scales displayed! fig.tight_layout() # plt.show() plt.savefig("fig13.png", dpi=300)
Figure 14
from Bio import Phylo import matplotlib.pyplot as plt tree = Phylo.read("tree.newick", "newick") tip_distances = [tree.distance(tree.root, tip) for tip in tree.get_terminals()] max_tip_distance = max(tip_distances) if tip_distances else 0 halfway = max_tip_distance / 2 label_offset = 0.05 fig, ax = plt.subplots() Phylo.draw( tree, do_show=False, axes=ax, label_func=lambda clade: clade.name if clade.is_terminal() else None, ) ax.axvline(x=halfway, color="red") ax.axvline(x=max_tip_distance, color="blue") ax.text( halfway - label_offset, 0.98, f"halfway={halfway:.3g}", color="red", ha="right", va="top", transform=ax.get_xaxis_transform(), ) ax.text( max_tip_distance - label_offset, 0.90, f"last tip={max_tip_distance:.3g}", color="blue", ha="right", va="top", transform=ax.get_xaxis_transform(), ) ax.yaxis.set_ticks([]) ax.set_ylabel("") ax.set_xlabel("Branch length") fig.savefig("fig14.png", dpi=300)