ggplot2 gallery

- colorbrewer2
- DataVisProject: a catalogue of visualisation types
- ggplot2 extensions
- R Graph Gallery
- R spatial blog posts
Gallery
Figure 1
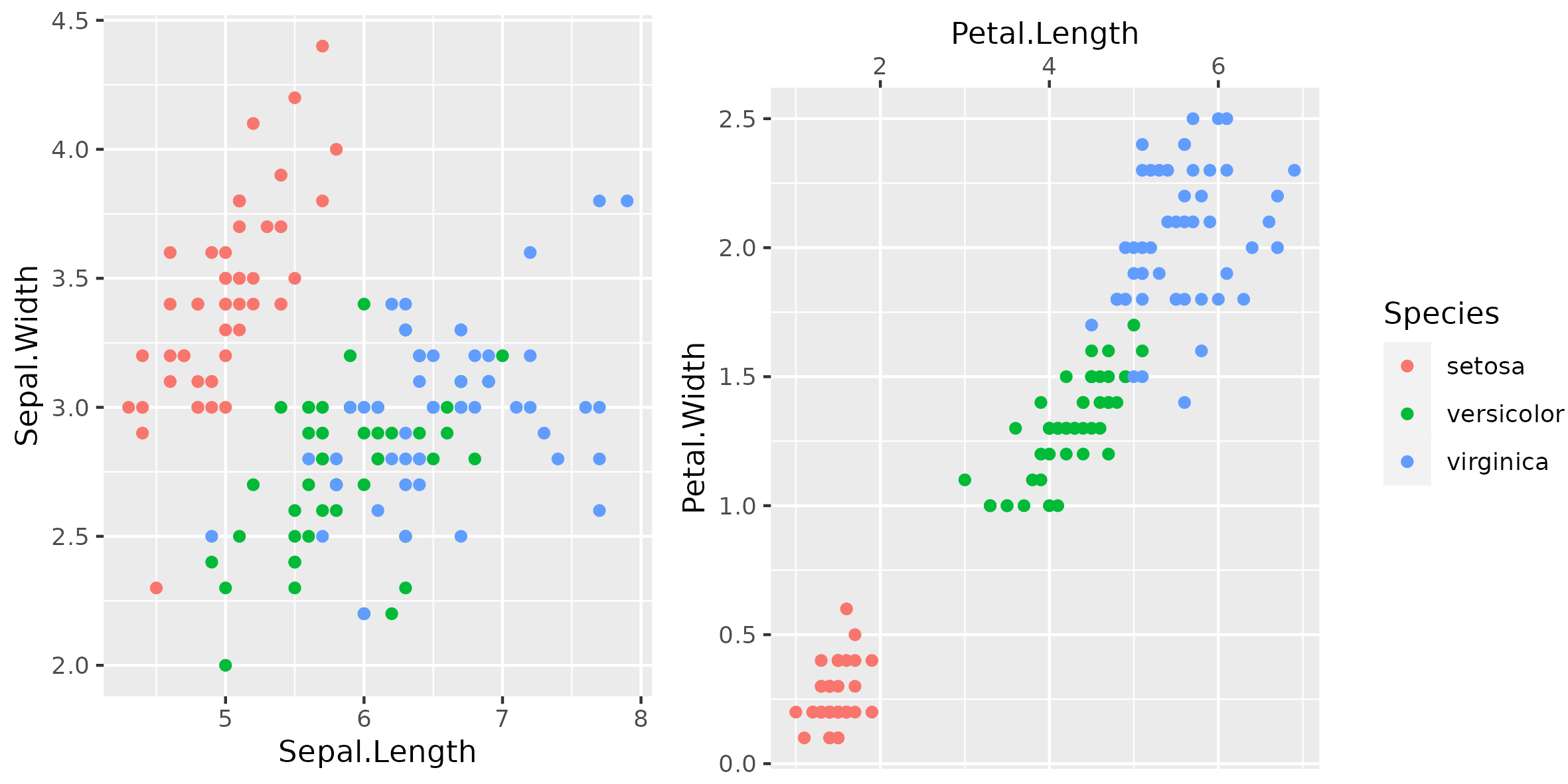
Keywords: cowplot, combining plots
library(ggplot2) library(cowplot) a <- ggplot() + geom_point(data = iris, mapping = aes(x = Sepal.Length, y = Sepal.Width, colour = Species)) b <- ggplot() + geom_point(data = iris, mapping = aes(x = Petal.Length, y = Petal.Width, colour = Species)) + scale_x_continuous(position = "top") legend <- get_legend(b + theme(legend.box.margin = margin(0, 0, 0, 12))) example_plot <- plot_grid(a + theme(legend.position = "none"), b + theme(legend.position = "none"), legend, ncol = 3, rel_widths = c(1, 1, 0.35)) ggsave("fig01.png", example_plot, width = 20, height = 10, units = "cm")
Figure 2
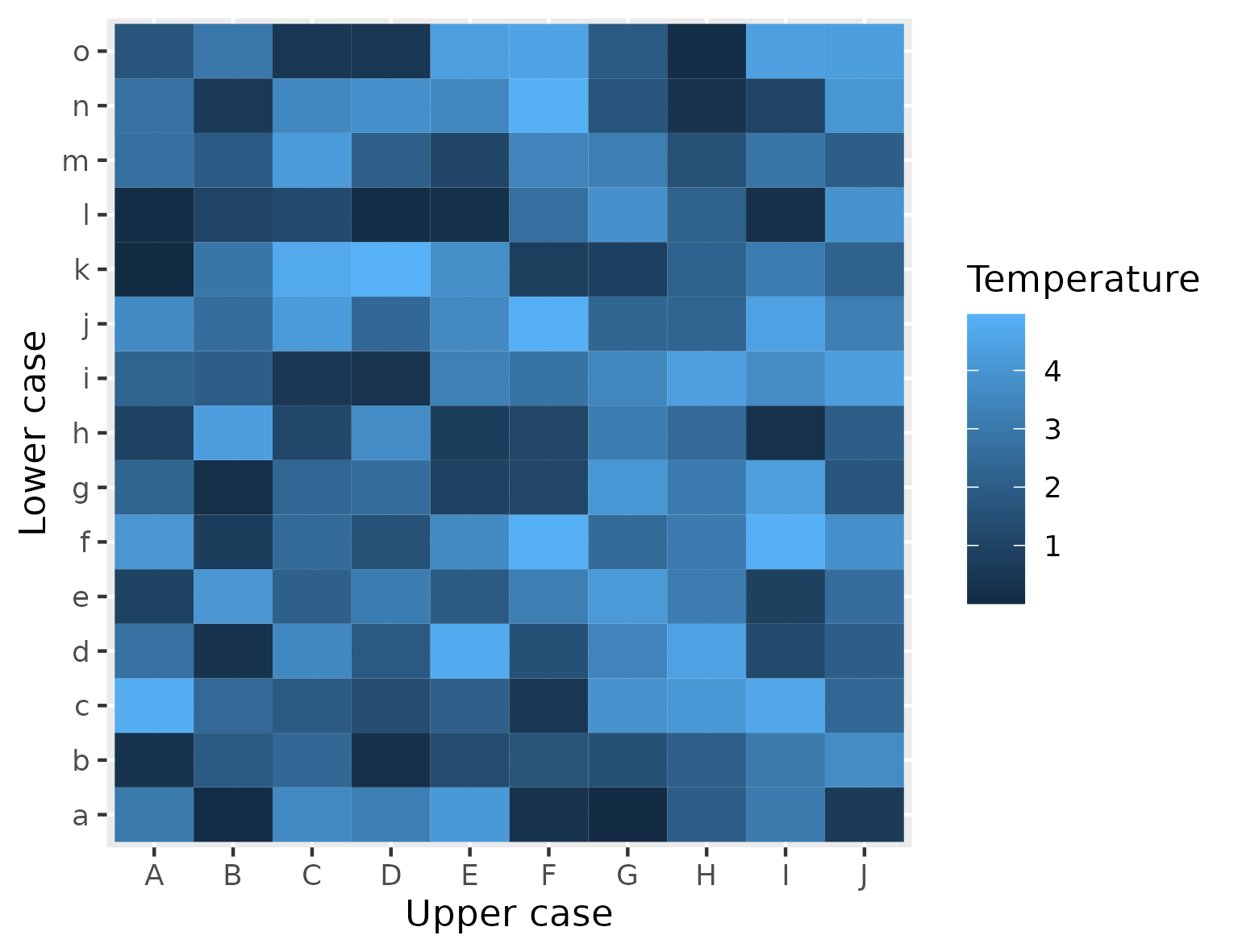
Keywords: heatmap, matrix, tile geometry
library(ggplot2) x <- LETTERS[1:10] y <- letters[1:15] plot_df <- expand.grid(x=x, y=y) plot_df$z <- runif(length(x) * length(y), 0, 5) example_plot <- ggplot() + geom_tile( data = plot_df, mapping = aes(x = x, y = y, fill = z) ) + labs( x = "Upper case", y = "Lower case", fill = "Temperature") theme_minimal() ggsave("fig02.png", example_plot, width = 13, height = 10, units = "cm")
Figure 3
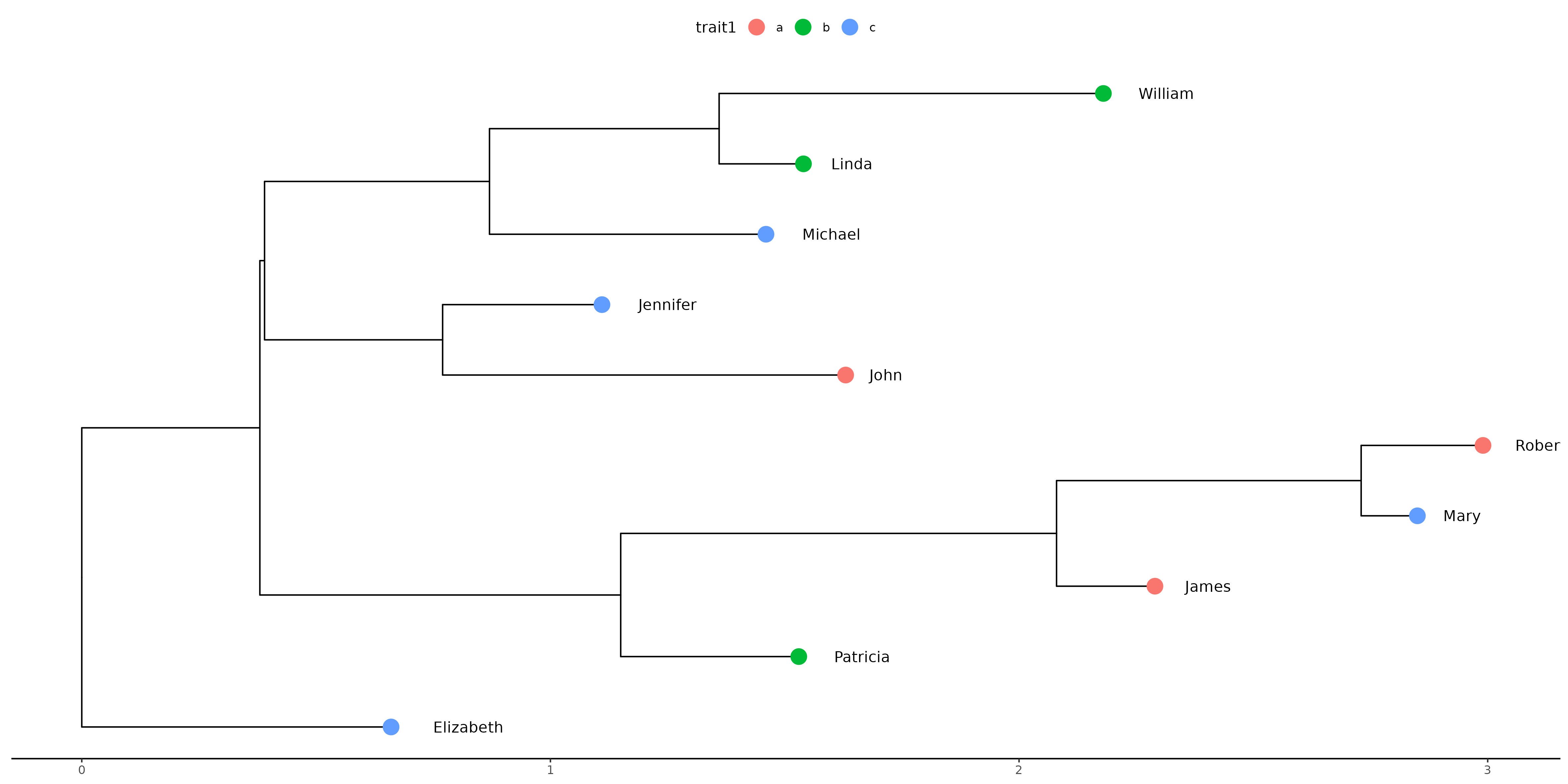
Keywords: tree, phylogeny, ggtree, newick
library(ggplot2) library(ggtree) library(treeio) library(tidytree) set.seed(1) newick_str <- "((((t7:0.21,(t2:0.12,t3:0.26):0.65):0.93,t8:0.38):0.77,((t1:0.86,t5:0.34):0.38,(t6:0.59,(t9:0.18,t10:0.82):0.49):0.48):0.01):0.38,t4:0.66);" my_phylo <- read.tree(text = newick_str) human_names <- c( "James", "Mary", "Robert", "Patricia", "John", "Jennifer", "Michael", "Linda", "William", "Elizabeth" ) annotations <- tibble( node = seq.int(length(my_phylo$tip.label)), trait1 = sample(letters[1:3], 10, replace = TRUE), trait2 = human_names ) my_tree <- treedata(phylo = my_phylo, data = annotations) example_plot <- ggplot(my_tree, mapping = aes(x, y)) + geom_tree() + geom_tippoint(mapping = aes(colour = trait1), size = 5) + geom_tiplab(mapping = aes(label = trait2), hjust = -0.5) + theme_tree2(legend.position='top') ggsave("fig03.png", example_plot, width = 40, height = 20, units = "cm")
Figure 4
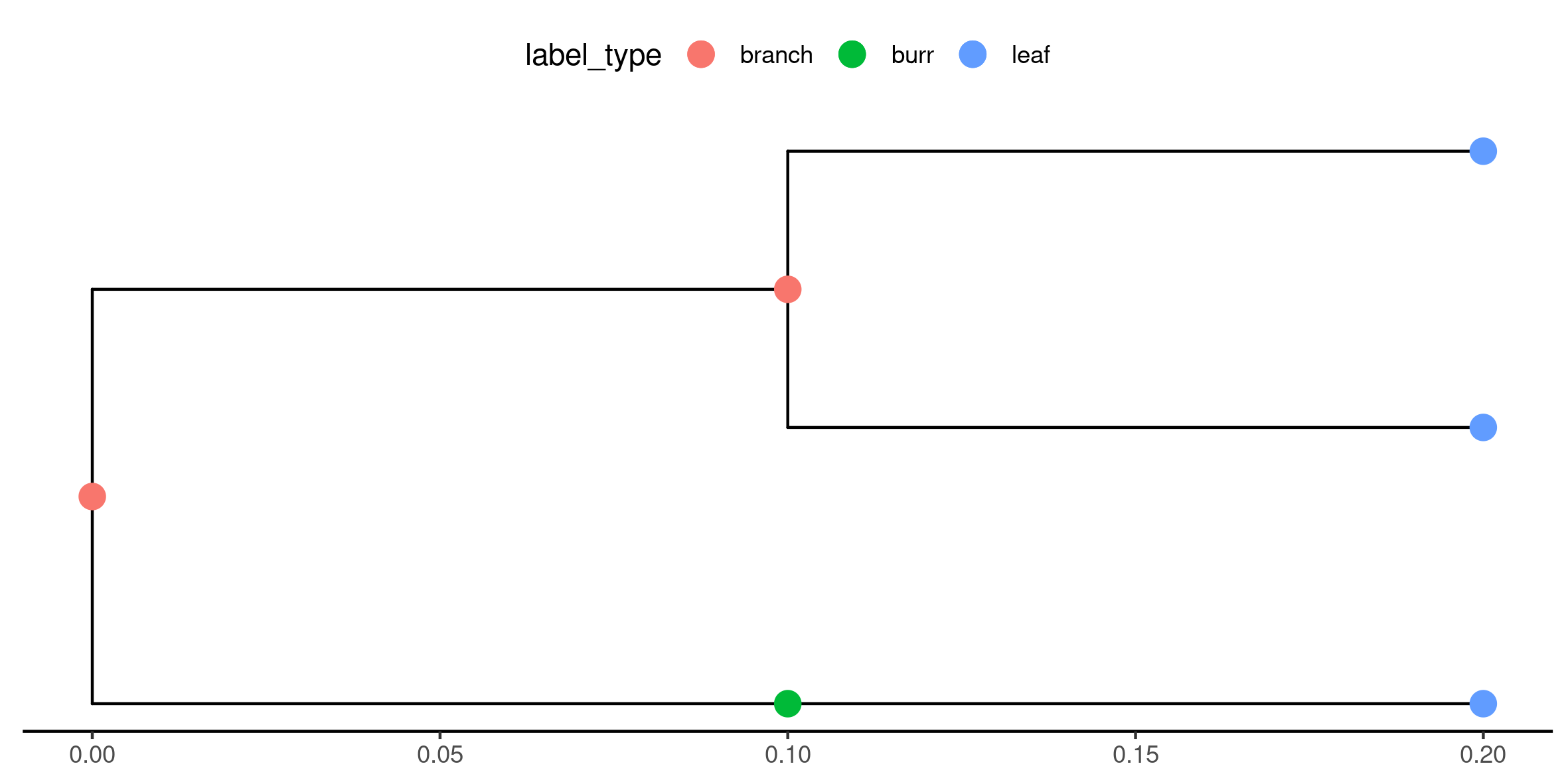
Keywords: tree, phylogeny, ggtree, newick, annotation
library(ggplot2) library(ggtree) library(treeio) library(tidytree) library(tibble) library(ape) set.seed(1) newick_str <- "((t1:0.1,t2:0.1):0.1,(t3:0.1)t4:0.1);" leaf_names <- c("t1", "t2", "t3") burr_names <- c("t4") label_type <- function(label) { ifelse( is.element(label, leaf_names), "leaf", ifelse( is.element(label, burr_names), "burr", "branch") ) } my_phylo_tbl <- as_tibble(read.tree(text = newick_str)) my_data_tbl <- tibble( node = my_phylo_tbl$node, label_type = label_type(my_phylo_tbl$label) ) my_td <- as.treedata( full_join(my_phylo_tbl, my_data_tbl, by = "node") ) example_plot <- ggplot(my_td, mapping = aes(x, y)) + geom_tree() + geom_tippoint(aes(colour = label_type), size = 4) + geom_nodepoint(aes(colour = label_type), size = 4) + theme_tree2(legend.position = "top") ggsave("fig04.png", example_plot, width = 20, height = 10, units = "cm")
Figure 5
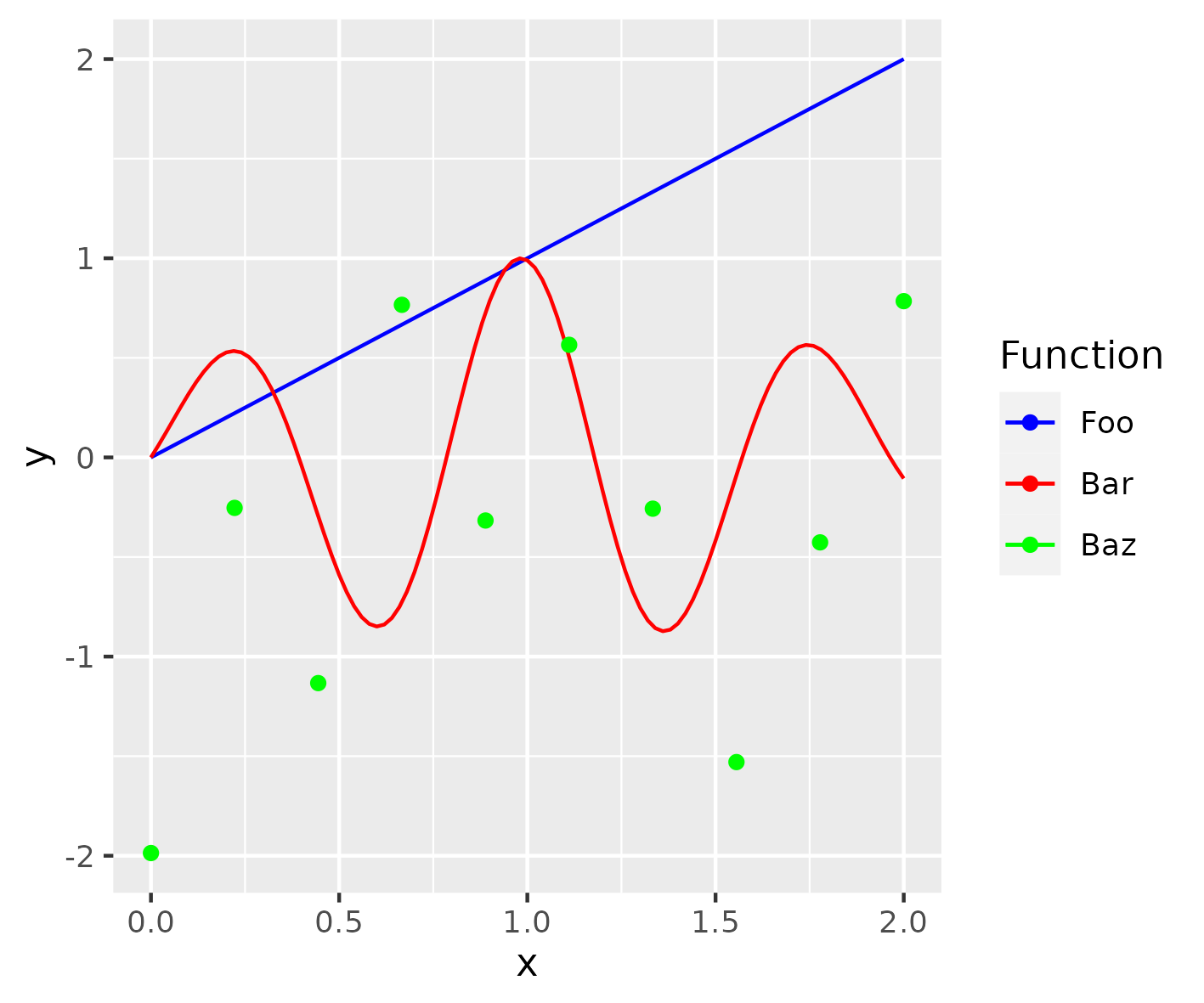
Keywords: function, curve, expression, colours
library(ggplot2) example_plot <- ggplot(data.frame(x = c(0, 2)), aes(x)) + stat_function( fun = identity, geom = "line", mapping = aes(colour = "Foo") ) + stat_function( fun = function(x) exp(-(x - 1)^2) * sin(x * 8), geom = "line", mapping = aes(colour = "Bar") ) + geom_point( data = data.frame( x = seq(0, 2, length = 10), y = rnorm(10) ), mapping = aes(colour = "Baz", y = y) ) + scale_colour_manual("Function", values = c("blue", "red", "green"), breaks = c("Foo", "Bar", "Baz") ) ggsave("fig05.png", example_plot, width = 12, height = 10, units = "cm")
Figure 6
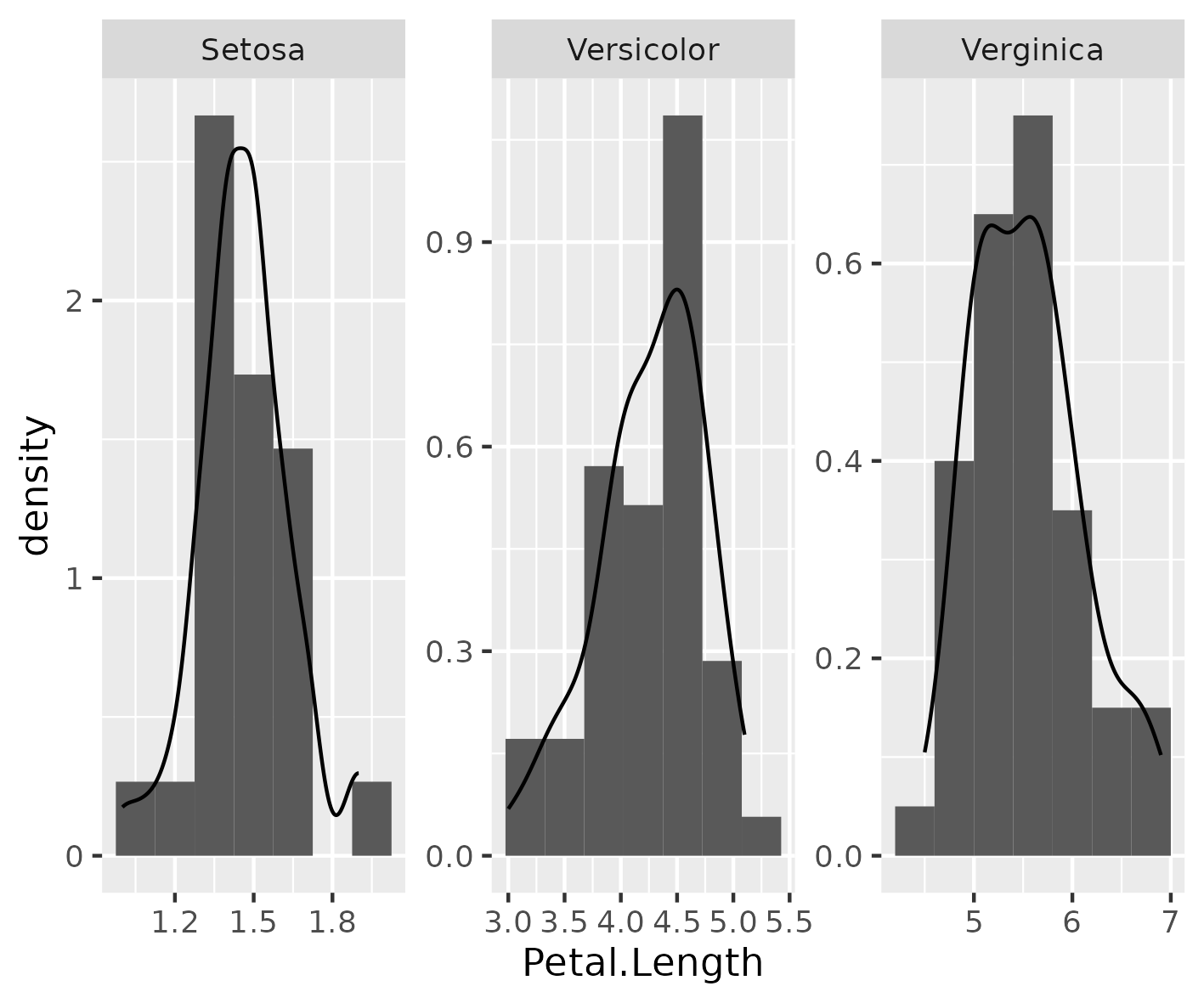
Keywords: facet labels
library(dplyr) library(ggplot2) plot_df <- iris %>% select(Petal.Length, Species) facet_labels <- c( setosa = "Setosa", versicolor = "Versicolor", virginica = "Verginica" ) example_plot <- ggplot(plot_df, aes(x = Petal.Length)) + geom_histogram( mapping = aes(y = ..density..), bins = 7 ) + geom_density() + facet_wrap(~Species, scales = "free", labeller = labeller(Species = facet_labels) ) ggsave("fig06.png", example_plot, width = 12, height = 10, units = "cm")
Figure 7
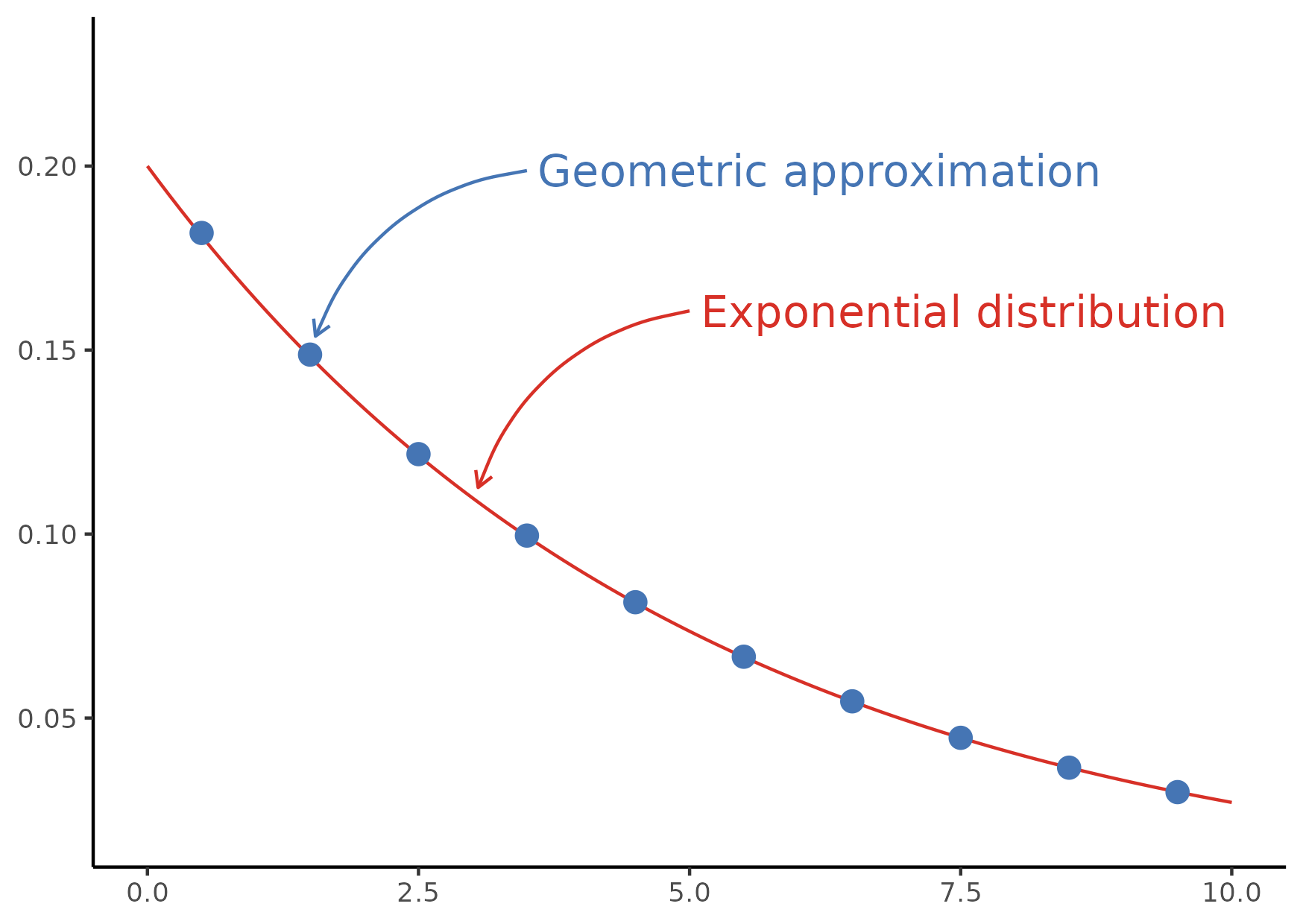
Keywords: annotations, arrows, colours
library(ggplot2) library(dplyr) ## Some hex codes to use as a colour scheme. These are from ## \code{https://colorbrewer2.org/?type=diverging&scheme=RdYlBu&n=7} my_colours <- list( truth = list(dark = "#d73027", light = "#fee090"), estimate = list(dark = "#4575b4", light = "#e0f3f8") ) exp_rate <- 0.2 x_max <- 10 xs <- seq(from = 0, to = x_max, length = 100) pdf_xs <- dexp(x = xs, rate = exp_rate) delta <- 1.0 p_hat <- 2 * exp_rate * delta / (exp_rate * delta + 2) n_max <- ceiling(x_max / delta - 0.5) ns <- seq.int(from = 0, to = n_max) ys <- delta * (ns + 0.5) pmf_ys <- dgeom(x = ns, prob = p_hat) my_fig <- ggplot() + geom_line( data.frame(x = xs, y = pdf_xs), mapping = aes(x = x, y = y), colour = my_colours$truth$dark ) + geom_point( filter(data.frame(x = ys, y = pmf_ys), x < x_max), mapping = aes(x = x, y = y), colour = my_colours$estimate$dark, size = 3 ) + annotate( geom = "curve", xend = ys[2] + 0.05, yend = pmf_ys[2] + 0.005, x = ys[2] + 2, y = pmf_ys[2] + 0.05, curvature = 0.3, arrow = arrow(length = unit(2, "mm")), colour = my_colours$estimate$dark ) + annotate( geom = "text", x = ys[2] + 2 + 0.10, y = pmf_ys[2] + 0.05, label = "Geometric approximation", hjust = "left", colour = my_colours$estimate$dark, size = 5 ) + annotate( geom = "curve", xend = 0.5*(ys[3] + ys[4]) + 0.05, yend = 0.5*(pmf_ys[3] + pmf_ys[4]) + 0.002, x = 0.5*(ys[3] + ys[4]) + 2, y = 0.5*(pmf_ys[3] + pmf_ys[4]) + 0.05, curvature = 0.3, arrow = arrow(length = unit(2, "mm")), colour = my_colours$truth$dark ) + annotate( geom = "text", x = 0.5*(ys[3] + ys[4]) + 2 + 0.10, y = 0.5*(pmf_ys[3] + pmf_ys[4]) + 0.05, label = "Exponential distribution", hjust = "left", colour = my_colours$truth$dark, size = 5 ) + ylim(c(0.02, 0.23)) + theme_classic() + theme(axis.title = element_blank()) ggsave(filename = "fig07.png", plot = my_fig, height = 10.5, width = 14.8, units = "cm")
Figure 8
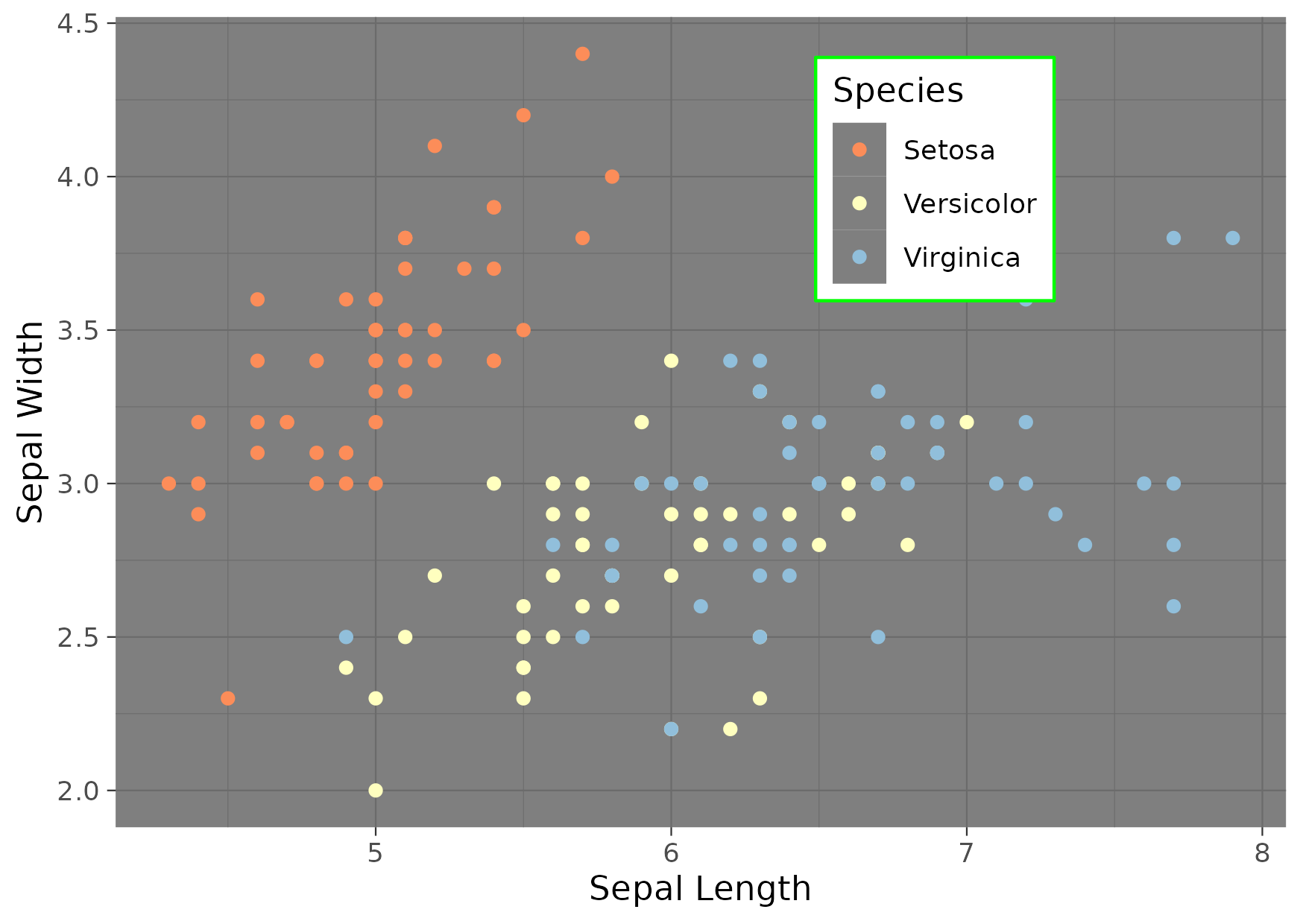
Keywords: Scales, colours, legends
library(ggplot2) hex_cols <- c('#fc8d59','#ffffbf','#91bfdb') gg <- ggplot() + geom_point( data = iris, mapping = aes(x = Sepal.Length, y = Sepal.Width, colour = Species) ) + scale_color_manual( values = hex_cols, breaks = c("setosa", "versicolor", "virginica"), labels = c("Setosa", "Versicolor", "Virginica") ) + labs( x = "Sepal Length", y = "Sepal Width" ) + theme_dark() + theme( legend.background = element_rect(fill = "white", colour = "green"), legend.position = c(0.7, 0.8) ) ggsave("fig08.png", gg, height = 10.5, width = 14.8, units = "cm")
Figure 0
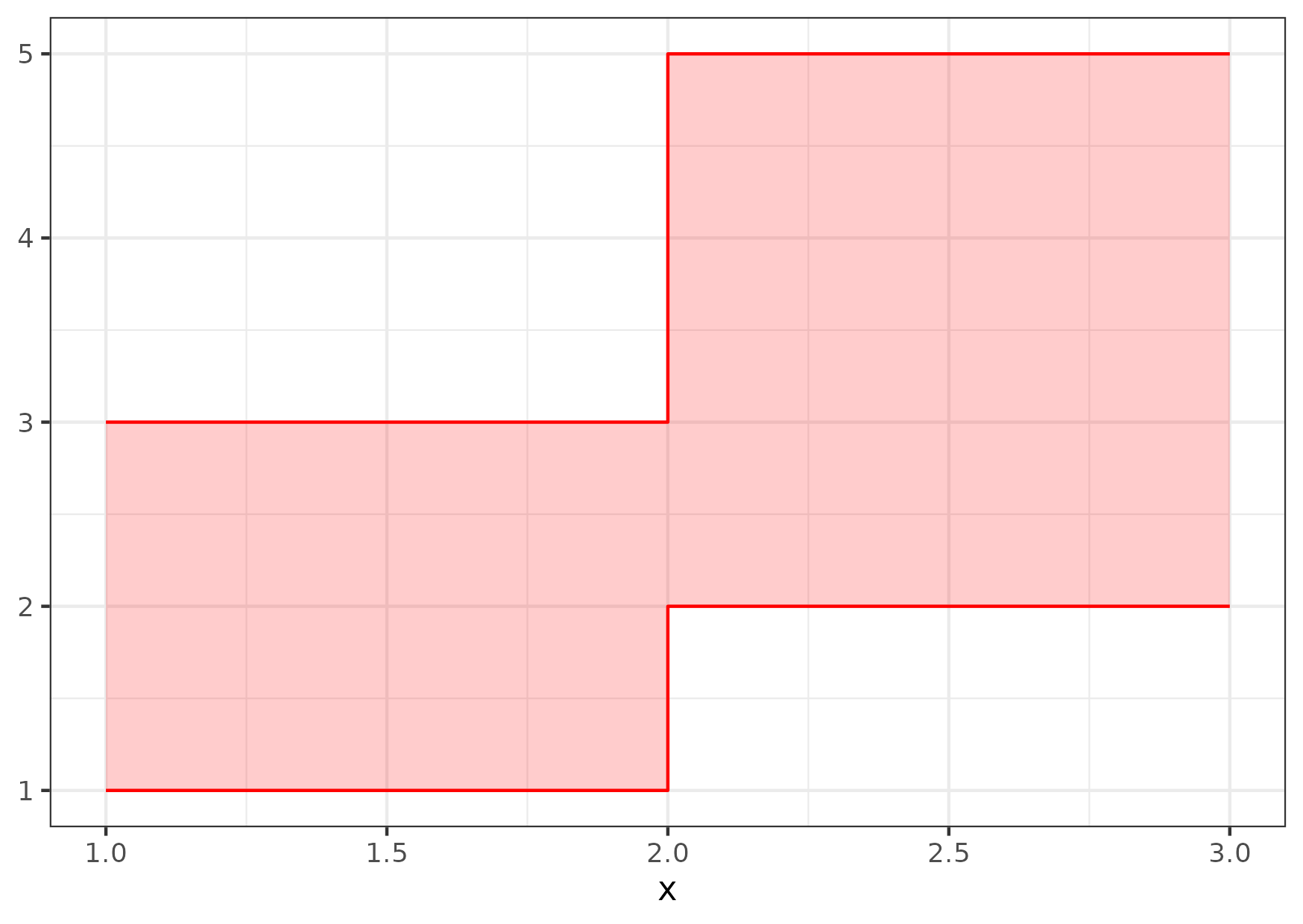
Keywords: geometries, ribbons, step, helper
library(ggplot2) #' Make a data frame for a ribbon step plot #' #' @param xs x values #' @param ymins y minimum values #' @param ymaxs y maximum values #' #' @examples #' plot_df <- make_ribbonstep_df(xs = c(1, 2, 3), #' ymins = c(1, 2), #' ymaxs = c(3, 5)) #' print(plot_df) #' ggplot() + #' geom_ribbon(data = plot_df, #' mapping = aes(x = x, ymin = ymin, ymax = ymax), #' color = "red", #' fill = "red", #' alpha = 0.2) + #' theme_minimal() make_ribbonstep_df <- function(xs, ymins, ymaxs) { stopifnot(length(xs) == length(ymins) + 1) stopifnot(length(ymins) == length(ymaxs)) middle_xs <- xs[-c(1, length(xs))] data.frame(x = c(xs[1], rep(middle_xs, each = 2), xs[length(xs)]), ymin = rep(ymins, each = 2), ymax = rep(ymaxs, each = 2)) } plot_df <- make_ribbonstep_df(xs = c(1, 2, 3), ymins = c(1, 2), ymaxs = c(3, 5)) gg <- ggplot() + geom_ribbon(data = plot_df, mapping = aes(x = x, ymin = ymin, ymax = ymax), colour = "red", fill = "red", alpha = 0.2) + theme_bw() ggsave("fig09.png", gg, height = 10.5, width = 14.8, units = "cm")
Figure 10
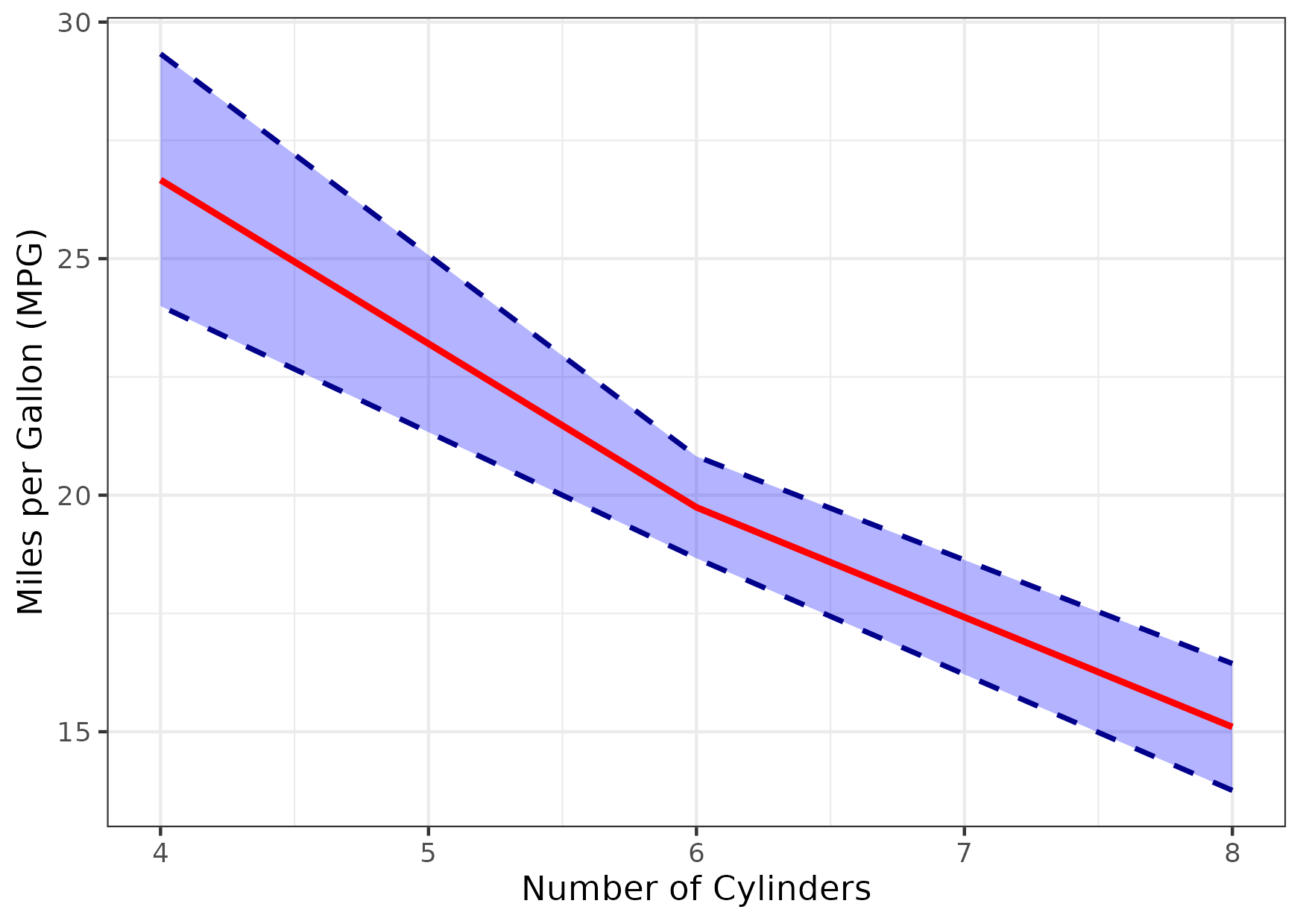
Keywords: geometries, ribbons, step, helper
library(ggplot2) library(dplyr) mtcars_ext <- mtcars |> group_by(cyl) |> summarize(mean_mpg = mean(mpg), sd_mpg = sd(mpg), lower_mpg = mean_mpg - 1.96 * sd_mpg / sqrt(n()), upper_mpg = mean_mpg + 1.96 * sd_mpg / sqrt(n())) |> ungroup() gg <- ggplot(mtcars_ext, aes(x = cyl)) + geom_ribbon(aes(ymin = lower_mpg, ymax = upper_mpg), fill = "blue", alpha = 0.3, color = "darkblue", linetype = "dashed", size = 0.8, show.legend = FALSE) + geom_line(aes(y = mean_mpg), color = "red", linewidth = 1, linetype = "solid", show.legend = FALSE) + labs(x = "Number of Cylinders", y = "Miles per Gallon (MPG)") + theme_bw() ggsave("fig10.png", gg, height = 10.5, width = 14.8, units = "cm")
Figure 11
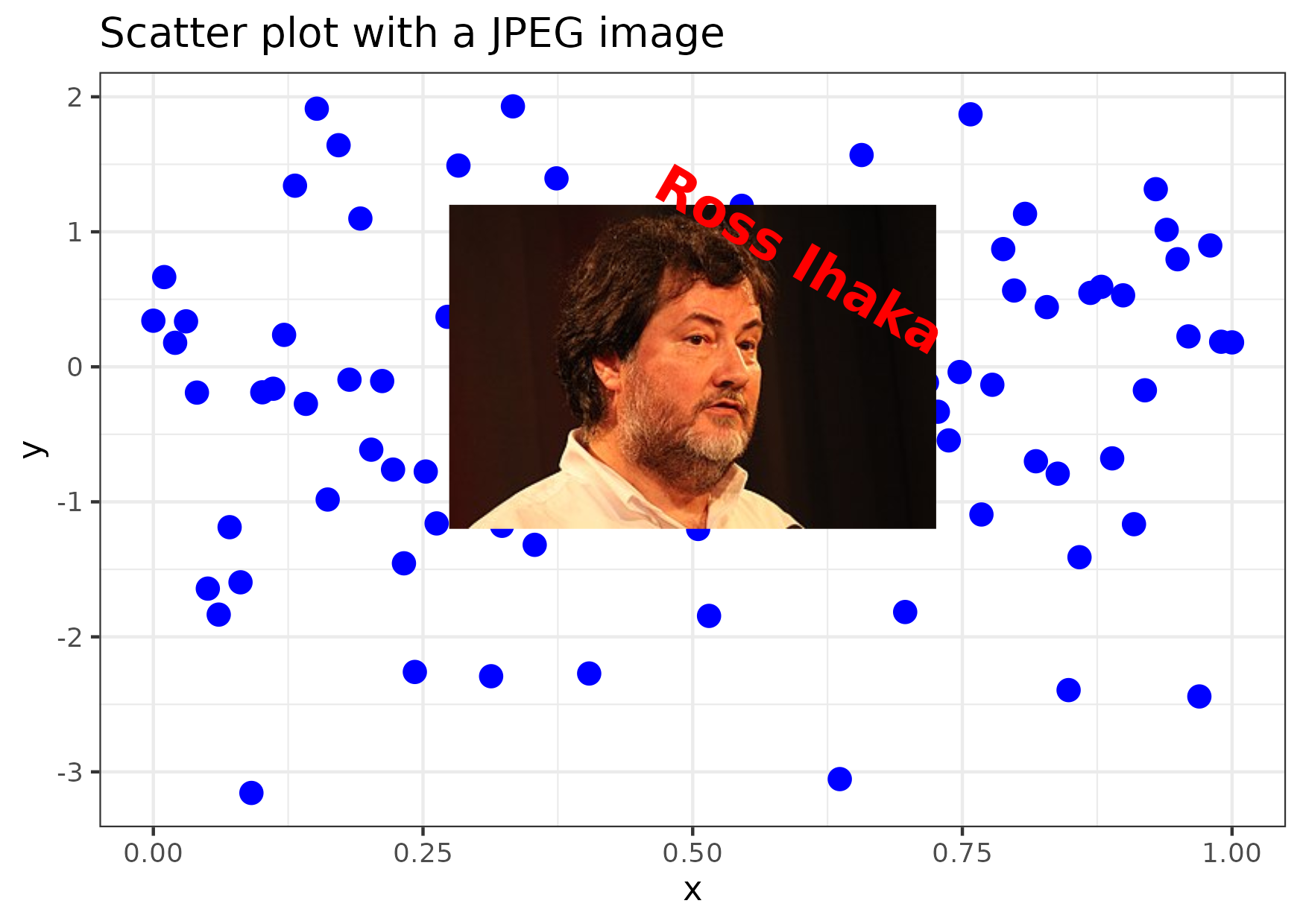
Keywords: images, annotation, pictures, png, jpeg
library(ggplot2) library(grid) ## Alternatively ## ## library(png) ## img_path <- "path/to/your/image.png" ## img <- readPNG(img_path) ## img_raster <- rasterGrob(img, interpolate = TRUE) library(jpeg) img_path <- "ross-ihaka.jpg" img <- readJPEG(img_path) img_raster <- rasterGrob(img, interpolate = TRUE) df <- data.frame(x = seq(from = 0, to = 1, length = 100), y = rnorm(100)) gg <- ggplot(df, aes(x, y)) + geom_point(color = "blue", size = 3) + annotation_custom( img_raster, xmin = 0.1, xmax = 0.9, ymin = -1.2, ymax = 1.2 ) + annotate( geom = "text", x = 0.6, y = 0.8, label = "Ross Ihaka", colour = "red", size = 6, fontface = "bold", angle = -30 ) + labs(title = "Scatter plot with a JPEG image") + theme_bw() ggsave("fig11.png", gg, height = 10.5, width = 14.8, units = "cm")
Figure 12
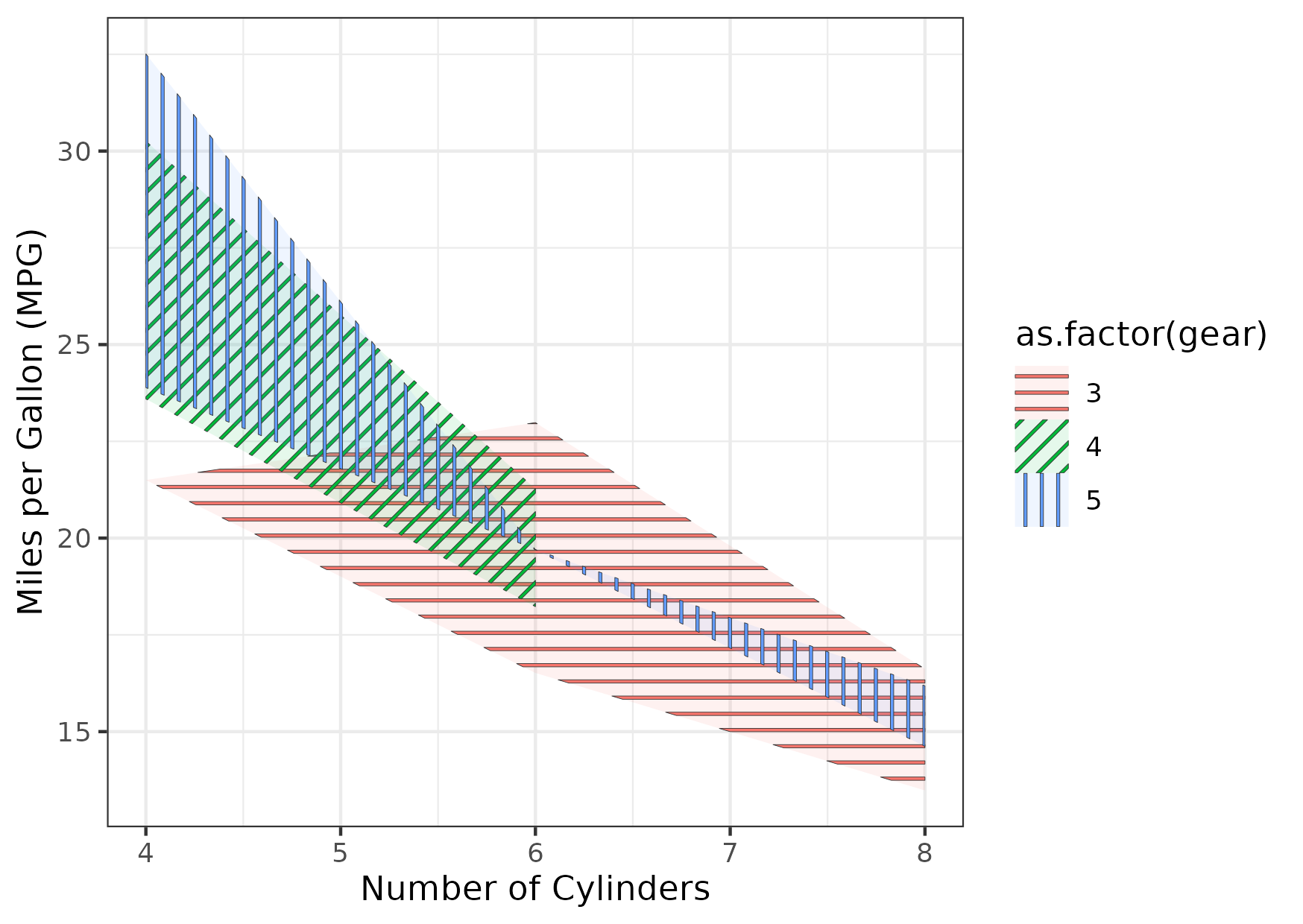
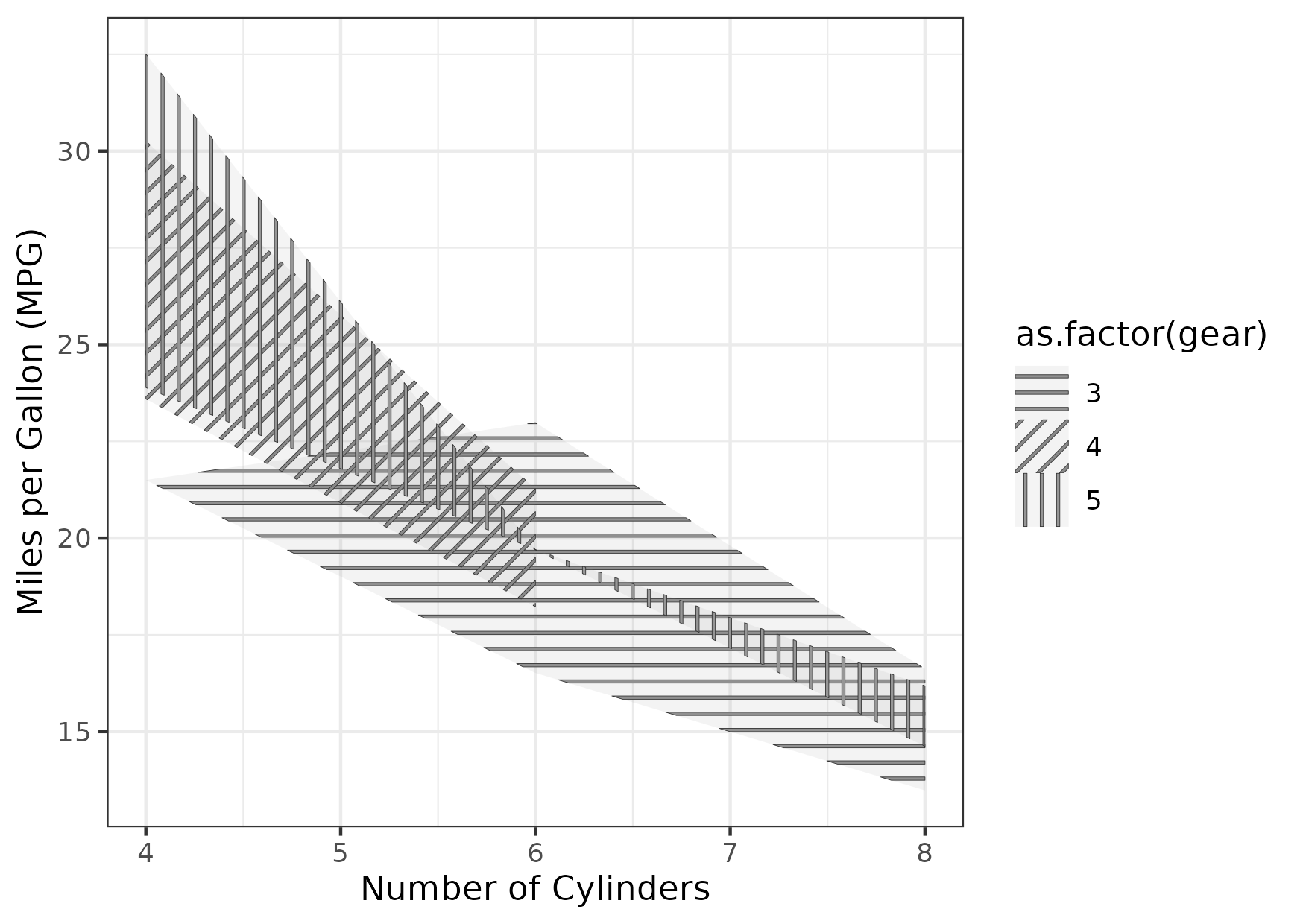
Keywords: patterns, black and white, grayscale, redundant
library(ggplot2) library(dplyr) library(tidyr) library(ggpattern) mtcars_ext <- mtcars |> group_by(cyl, gear) |> summarize(mean_mpg = mean(mpg), sd_mpg = ifelse(is.na(sd(mpg)), 0, sd(mpg)), lower_mpg = mean_mpg - 1.96 * sd_mpg / sqrt(n()), upper_mpg = mean_mpg + 1.96 * sd_mpg / sqrt(n())) |> ungroup() gg <- ggplot() + geom_ribbon_pattern( data = mtcars_ext, mapping = aes(x = cyl, ymin = lower_mpg, ymax = upper_mpg, pattern_angle = as.factor(gear), pattern_fill = as.factor(gear), fill = as.factor(gear)), pattern = "stripe", pattern_spacing = 0.02, pattern_size = 0.1, alpha = 0.1) + labs(x = "Number of Cylinders", y = "Miles per Gallon (MPG)") + theme_bw() ggsave("fig12.png", gg, height = 10.5, width = 14.8, units = "cm")
Figure 13
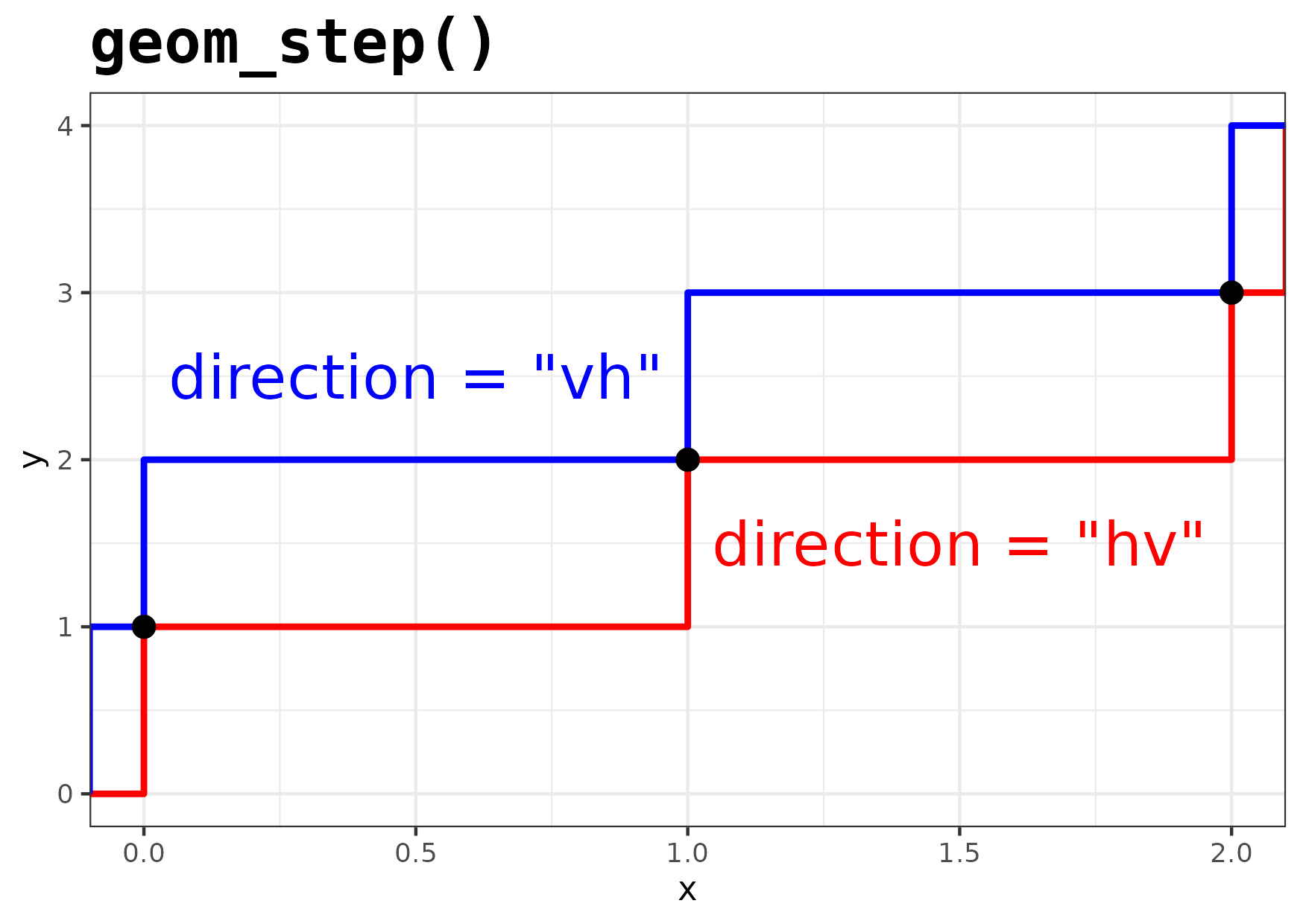
Keywords: step function, piece-wise constant, horizontal
library(ggplot2) library(dplyr) plot_df <- data.frame(x = c(-Inf, 0, 1, 2, Inf), y = c(0, 1, 2, 3, 4)) text_size <- 7 gg <- ggplot() + geom_step( data = plot_df, mapping = aes(x = x, y = y), direction = "hv", colour = "red", size = 1) + geom_step( data = plot_df, mapping = aes(x = x, y = y), direction = "vh", colour = "blue", size = 1) + geom_point( data = filter(plot_df, is.finite(x)), mapping = aes(x = x, y = y), size = 3) + geom_text( mapping = aes(x = 0.5, y = 2.5, label = "direction = \"vh\""), size = text_size, colour = "blue" ) + geom_text( mapping = aes(x = 1.5, y = 1.5, label = "direction = \"hv\""), size = text_size, colour = "red" ) + labs( title = "geom_step()" ) + theme_bw() + ## Use a fixed with font for the title text theme(plot.title = element_text(size = 20, family = "mono", face = "bold")) ggsave("fig13.png", gg, height = 10.5, width = 14.8, units = "cm")
Figure 14
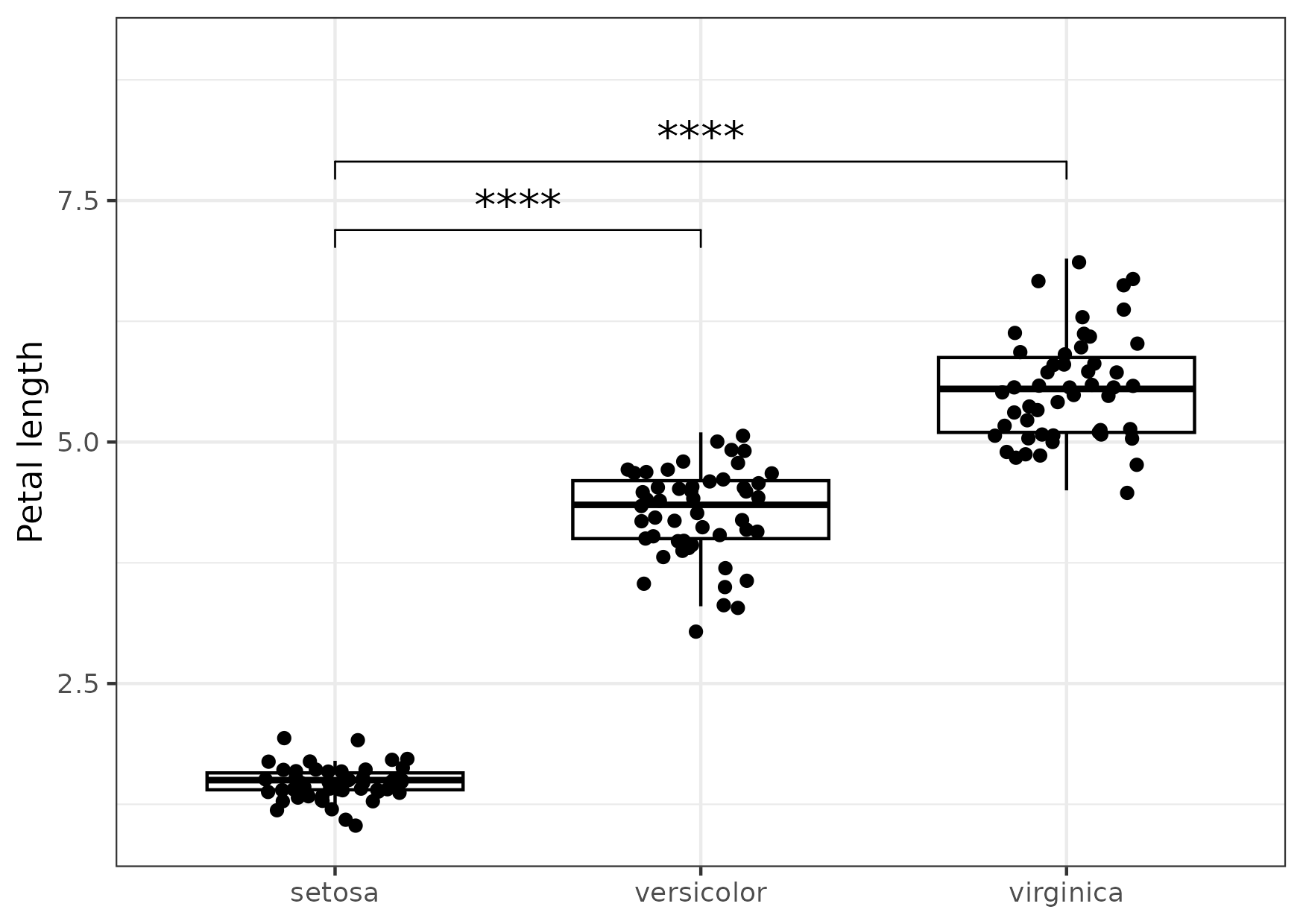
Keywords: ggpubr, significance, publication, comparison
library(ggplot2) library(dplyr) library(ggpubr) my_comparisons <- list(c("setosa", "versicolor"), c("setosa", "virginica")) gg <- ggboxplot(data = iris, x = "Species", y = "Petal.Length", add = "jitter") + stat_compare_means(comparisons = my_comparisons, label = "p.signif", size = c(5, 5)) + scale_y_continuous(limits = c(1, 9)) + labs(x = NULL, y = "Petal length") + theme_bw() ggsave("fig14.png", gg, height = 10.5, width = 14.8, units = "cm")
Colophon
if (!requireNamespace("BiocManager", quietly = TRUE)) install.packages("BiocManager") BiocManager::install("ggtree")